Genomics
 Genomics is the study of individual genomes, both at the sequence level and at the structural level. Technological advances have made genomic approaches critical for understanding basic biological processes, so that genomic research now impacts all areas of life science research.
Genomics is the study of individual genomes, both at the sequence level and at the structural level. Technological advances have made genomic approaches critical for understanding basic biological processes, so that genomic research now impacts all areas of life science research.
The CSHL Genomics Program includes faculty working across disciplines and research areas. Their main interests are genomic organization, structural variation of the human genome as related to disease, computational genomics, small RNA biology, transcriptional modeling, and sequencing technology. Genomics research at CSHL benefits from state-of-the-art technology and innovative software development, with researchers both on the main CSHL campus and at the nearby CSHL Woodbury Genome Center.
Broadly, genomics research falls into three categories with significant overlap: Cognitive Genomics, Cancer Genomics and Plant Genomics. In addition, Genomics researchers works closely with researchers in other CSHL programs, particularly the Simon Center for Quantitative Biology.
CSHL is one of the founding members of the New York Genome Center, an independent, non-profit organization that is leveraging the collaborative resources of leading medical and research institutions to transform medical research and clinical care in New York.
Cocktails & Chromosomes: Heresy in genetics
June 17, 2025
Imagine everything you learned about DNA wasn’t quite wrong, but was only a fraction of the truth. Now, get more of the whole story.
Exposomics: Holistic health without the snake oil
June 12, 2025
After gathering at Cold Spring Harbor Laboratory’s Banbury Center, the field’s leading minds issue a rallying cry heard across biology and medicine.
CSHL and global collaborators map Solanum pan-genome
March 5, 2025
Landmark study carves a path for plant breeders to improve crops across the globe and expand the variety of produce available in grocery stores.
Corn’s ancient ancestors are calling
February 6, 2025
Cold Spring Harbor Laboratory Professors Thomas Gingeras and Rob Martienssen have launched a new genomic encyclopedia called MaizeCODE.
Big clues emerge in small RNA mystery
December 16, 2024
How does the body separate “self” from “nonself?” CSHL Professor Rob Martienssen uncovers fascinating new leads in plant pollen and mouse sperm.
Cocktails & Chromosomes: Long-lived creatures of the night
November 19, 2024
Bat genes may hold the keys to aging, immunity, and cancer. Find out how, as we relive a special Halloween night event with CSHL’s Dick McCombie.
New DNALC program aims to improve pandemic response
November 4, 2024
The federally funded initiative will train undergraduate teachers in disease-tracking techniques that could help prevent future pandemics.
At the Lab Season 1 Research Rewind: Genetics
October 22, 2024
It’s the code for all life on Earth. This week At the Lab, we’re hacking it with the help of Cold Spring Harbor Laboratory’s geneticists.
At the Lab Episode 26: The golden grail of genomics
October 1, 2024
For our Season 1 finale, we invite you to step inside EN-TEx, a catalog of more than one million genomic variants.
At the Lab Episode 25: How maize became corn
September 24, 2024
Cold Spring Harbor Laboratory solves a plant biology mystery some 4,000 years in the making. The implications may go far beyond vegetables.
CSHL grad student wins International Birnstiel Award
September 23, 2024
Shushan Toneyan won the award for her thesis research in CSHL’s Koo lab. Toneyan is the co-creator of CREME, an AI-powered virtual laboratory.
Is CREME AI’s answer to CRISPR?
September 16, 2024
CREME, the latest AI toolkit from CSHL, is a virtual laboratory that may help scientists find new therapeutic targets in the genome.
Corn’s ‘missing link’
August 7, 2024
CSHL researchers have discovered a biological mechanism that may explain how corn spread so rapidly across the Americas 4,000 years ago.
Every rose has its thorns … or does it?
August 1, 2024
CSHL biologists have found a way to remove prickles from plants genetically. Their discovery could change the way we think about evolution in general.
At the Lab Episode 17: AI SQUID
July 30, 2024
Tune in to this week’s podcast to hear about the latest artificial intelligence model coming out of Cold Spring Harbor Laboratory.
Welcome to the summer of sequencing
July 17, 2024
CSHL’s DNA Learning Center and collaborators traveled cross-country to show educators the power of portable DNA sequencing.
SQUID pries open AI black box
June 21, 2024
CSHL’s Koo and Kinney labs have built a tool to suss out how AI analyzes the genome. What sets it apart? Decades of quantitative genetics knowledge.
Autism genetics: The faces behind the data
May 16, 2024
CSHL research on autism involves massive databases with thousands of genomes. Meet a few of the brave individuals who help make this work possible.
At the Lab Episode 4: DNA sequencing for all
April 23, 2024
CSHL Dolan DNA Learning Center’s Jason Williams teaches us about a portable DNA sequencer that may turn the tide for genetics education.
At the Lab Episode 3: Autism Awareness Month
April 16, 2024
It’s Autism Awareness Month! CSHL Professor Ivan Iossofiv shares what researchers know about autism so far and how they plan to uncover its origins.
CSHL’s Thomas Gingeras awarded $2 million NSF grant
April 3, 2024
Climate change threatens crops with acidic soils and aluminum toxicity. Gingeras leads an international team tackling this problem head-on.
From plant genomics to a bioscience revolution
March 25, 2024
CSHL played a lead role in mapping the first plant genome. Today, that breakthrough fuels a whole new understanding of life on Earth.
Cocktails & Chromosomes: Sequencing suds
February 15, 2024
CSHL’s series of fun, interactive science talks returns to Industry bar in Huntington, NY, with a demonstration of today’s DNA sequencing technology.
Genome sequencing for kids, one LEGO at a time
February 13, 2024
The CSHL DNA Learning Center’s LEGO DNA sequencer makes learning about the genome fun and easy for students of all ages, even kindergarteners.
From cancer genetics to cancer treatments
January 18, 2024
One cancer gene, one cancer genome, two Cold Spring Harbor Laboratory discoveries that helped shape the face of modern cancer medicine.
Joshua-Tor named CSHL Director of Research
January 2, 2024
The Cold Spring Harbor Laboratory professor and HHMI investigator steps into her new role effective January 2, 2024.
Inside CSHL’s Single-Cell Biology Facility
December 19, 2023
A decade ago, sequencing individual cells was a luxury. Today, it’s critical for biology research. Meet the scientists making it happen at CSHL.
Genome sequencing goes mobile at DNALC
August 31, 2023
New collaboration between CSHL’s DNA Learning Center and Oxford Nanopore Technologies puts DNA sequencing in the palms of students’ hands.
Where did our DNA come from?
August 22, 2023
Half the human genome isn’t quite human. CSHL’s Andrea Schorn gives us the inside scoop on how our DNA turned ancient viruses into essential allies.
DNA Learning Center teaches the teachers
August 11, 2023
Local educators learn the ropes of genetics and bioinformatics research so they can bring it back to their students and the public.
The digital dark matter clouding AI
June 5, 2023
Scientists have unknowingly encountered mysterious noise while using AI to decipher our genetic code. CSHL has found a way to cut through the fog.
The evolution of autism research
May 25, 2023
The conversation around autism has evolved over the past two decades. So has CSHL research. This retrospective shows how we’ve helped move the needle.
Autism in the family tree
May 23, 2023
CSHL scientists have studied the genetics of autism across hundreds of family trees. This animated video shows what they’ve found.
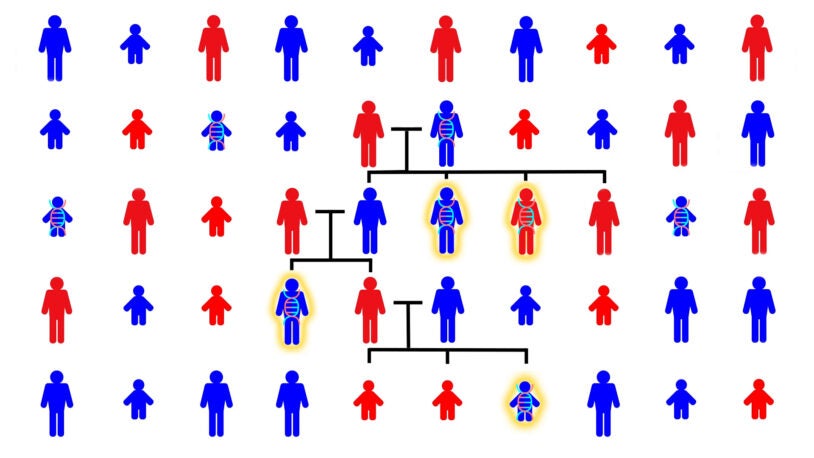
Siblings with autism share more of dad’s genome, not mom’s
May 22, 2023
CSHL study of more than 6,000 volunteer families overturns a long-held assumption about the genetic origins of autism spectrum disorder.
AI training: A backward cat pic is still a cat pic
May 4, 2023
This basic rule of thumb is helping CSHL’s quantitative biologists train AI to get a better read of the human genome.
Genome analysis just got personal
March 30, 2023
A new algorithm created by CSHL researchers can help predict the health effects of millions of genetic variants found within a single person’s genome.
Finding the right AI for you
December 5, 2022
AI’s popularity has reached a point where there are too many options. How do you know which AI is right for you? CSHL scientists have a solution.
New CSHL website brings together sorghum researchers
May 9, 2022
Plant researchers and breeders are now using a website created by CSHL to get the latest intel on sorghum crop research.
The race to protect sweet corn
April 22, 2022
Breeding a variety that can withstand disease and taste better too
For plant geneticists, some genes are double the trouble
March 28, 2022
It pays to check whether genetic tweaks that improve one kind of crop could get foiled by backup genes in a different crop.
Defining cells across diverse ancestries
November 17, 2021
CSHL Fellow Semir Beyaz is leading a multi-institutional effort to explore how genetic ancestry influences health and disease in the uterus.
CSHL Fellow Hannah Meyer wins UK Biobank researcher award
November 17, 2021
The UK Biobank recognized CSHL Fellow Hannah Meyer’s scientific achievements in understanding the inner workings of the human heart.
The secret history of corn is revealed in its genome
August 5, 2021
For the first time, scientists have assembled in-depth maps of dozens of corn genomes, filling in gaps related to key agricultural traits.
DNA Learning Center provides hands-on science at home
May 26, 2021
During the pandemic educators engaged with students virtually. The DNALC is preparing for live instruction at a new center in Brooklyn, NY.
Making AI algorithms show their work
May 13, 2021
AI machines are often better than humans at discerning patterns. CSHL researchers developed a way to find out why.
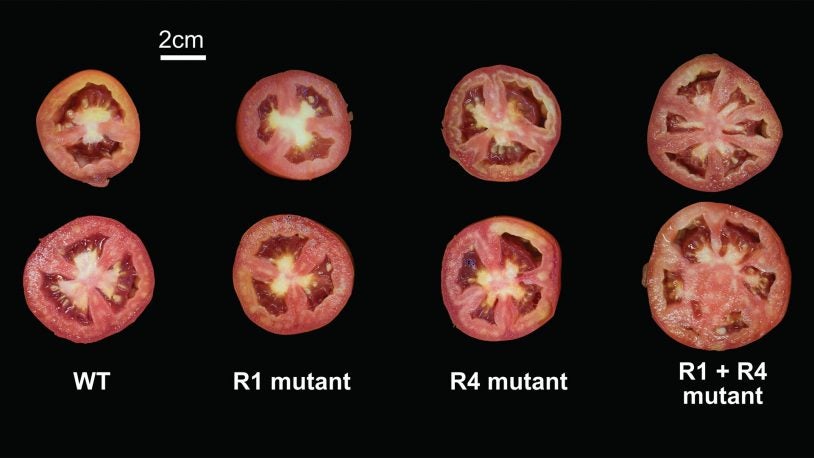
For tomato genes, one plus one doesn’t always make two
April 12, 2021
CSHL scientists made a series of mutations in tomatoes to see how they interact with one other. Some combinations yield larger changes than expected.
WOX9: A jack of all trades
March 4, 2021
A single gene can have many functions across different plant genomes. Changing the gene’s regulatory region can change the traits it produces.
The complete human genome: A “book of life”
February 24, 2021
Thirty years ago, scientists gathered at CSHL to plan the Human Genome Project (HGP). Today, the HGP provides the foundation for biological research.
Tweaking corn kernels with CRISPR
February 22, 2021
CRISPR genome editing can fast-forward the process of plant evolution. Researchers at CSHL are using the technique to increase kernel yield.
The world’s first DNA “tricorder” in your pocket
December 7, 2020
A new iPhone app, paired with a handheld sequencer, can recreate a genetics laboratory anywhere in the world.
Birds of a feather do flock together
November 17, 2020
Researchers found a genetic mechanism for how brand new species acquire distinct traits.
Human Nature documentary panel discussion
September 2, 2020
CRISPR experts, including Professors Zach Lippman, Jennifer Doudna, and Alta Charo discuss the gene-editing technology and its use.
CSHL hosts Human Nature documentary panel with CRISPR experts
September 2, 2020
CRISPR experts, including Professors Zach Lippman, Jennifer Doudna, and Alta Charo discuss the gene-editing technology and its use.
Martienssen named 2020 Royal Society winner
August 3, 2020
Professor and HHMI Investigator Rob Martienssen wins a 2020 Royal Society medal for his RNAi research.
ENCODE3: Interpreting the human and mouse genomes
July 29, 2020
Researchers report on 900,000 regulatory elements in our genomes that could influence our health.
The DNA tricks that gave us 100 different kinds of tomatoes
June 17, 2020
It takes 230,000 genetic differences to make 100 different varieties of tomatoes.
New faculty Jeff Boyd studies breast cancer genomics
March 26, 2020
Professor Jeff Boyd joins the CSHL faculty, studying the growth and spread of breast cancer.
DNA Learning Center takes New York
December 12, 2019
DNA Learning Center is expanding its educational reach into the greater New York area through the opening of two new centers.
Making sense of the genome…at last
December 6, 2019
Quantitative biologists like Cold Spring Harbor Laboratory’s Adam Siepel are finally making sense of the flood of data contained in the human genome.
The Lab partners with award-winning magazine
December 6, 2019
Nautilus, an award-winning science magazine, has partnered with CSHL to bring the story of the lab’s scientists and research to a brand-new audience.
Researchers double sorghum grain number to improve food supply
October 30, 2019
A set of hormone-controlling genes may be the key to doubling grain number in sorghum plants.
Seeking better treatment for ALS, Lou Gehrig’s disease
October 29, 2019
Researchers found that ‘jumping genes’ were de-silenced in post-mortem tissue samples of ALS patients.
Are smart robots a threat?
August 23, 2019
In a Q&A, Neuroscientist Anthony Zador explains how neuroscience can inform machine learning, and why he’s not worried about a robot apocalypse.
Creature feature quiz
July 31, 2019
Animals have been the muses behind major scientific breakthroughs for ages. See how much you know about the living organisms sharing the Earth with us.
Scientists teach old worms new tricks
June 23, 2019
Model organisms such as yeast, fruit flies, and worms have advanced the study of genomics, eukaryotic biology, and evolution.
An essay from the President: Biology for the planet
May 16, 2019
CSHL plant scientists are looking for solutions to the biggest questions in agriculture as environments are reshaped by climate change.
Cryptic mutation is cautionary tale for crop gene editing
May 6, 2019
Unexpected interactions between mutations can be a thorn in the side for plant breeders. Scientists unveil what drove one infamous “cryptic” mutation.
Andrea Schorn zooms in on small RNAs in the cell
April 29, 2019
Dr. Andrea Schorn joins CSHL faculty as research assistant professor. Her focus is exploring transposable elements in the genome of mammalian cells.
How does natural selection affect the genome?
December 18, 2018
Adam Siepel explains how natural selection can tell researchers how informative sifting through the complex human genome will be.
How much are we learning? Natural selection is science’s best critic
December 17, 2018
Researchers determine that natural selection and our evolutionary history may be the best guides for future research.
Base Pairs Episode 17.5: Special interview with Yaniv Erlich
October 16, 2018
We sat down with Dr. Yaniv Erlich, chief scientific officer at MyHeritage DNA, to get his unique perspective on the use of personal genetic info.
Alexander Dobin dives into genomic data
June 8, 2018
Alexander Dobin joins the faculty as its newest assistant professor, working on the computational side of genomics research
A science writer’s quest to understand heredity
May 30, 2018
LabDish spoke with science writer Carl Zimmer about what he learned about heredity as he zig-zagged through CSHL while writing his new book.
CRISPR-based system identifies important new drug targets in a deadly leukemia
March 8, 2018
CRISPR was used to find 2 new druggable targets in a deadly leukemia
The secret to tripling the number of grains in sorghum and perhaps other staple crops
February 26, 2018
A simple genetic modification can triple grain production in sorghum, a drought-tolerant plant that is an important source of food and animal feed.
More rice, please: 13 rice genomes reveal ways to keep up with ever-growing population
February 1, 2018
Rice provides 20% of calories consumed. As the population grows toward 9 billion and the climate shifts, we'll need to grow more rice in more places.
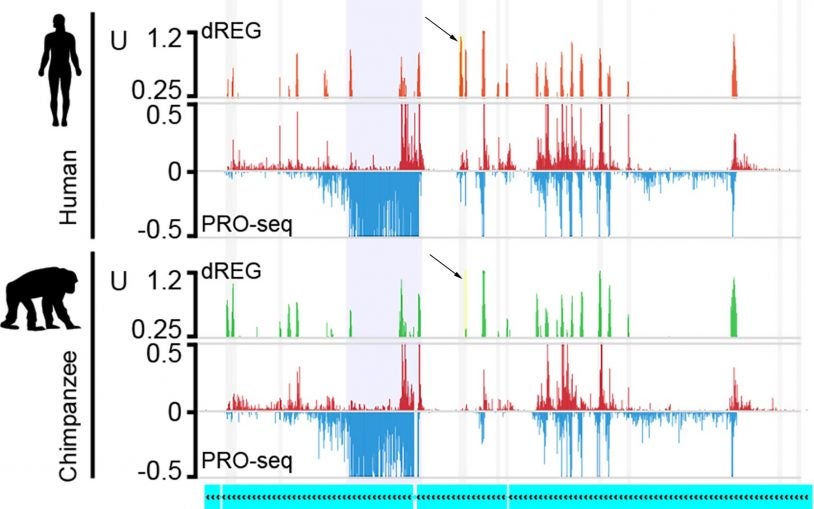
Evolving sets of gene regulators explain some of our differences from other primates
January 29, 2018
What makes us different from our primate relatives? Gene regulation is one important evolutionary factor
Hannon named director of Cancer Research UK Cambridge Institute
November 21, 2017
Dr. Greg Hannon has been named the new director of the Cancer Research UK Cambridge Institute, effective February 1, 2018.
What defends the genome when it’s “naked?”
July 20, 2017
Professor Rob Martienssen and postdoctoral researcher Andrea Schorn explain what defends the genome when it's "naked."
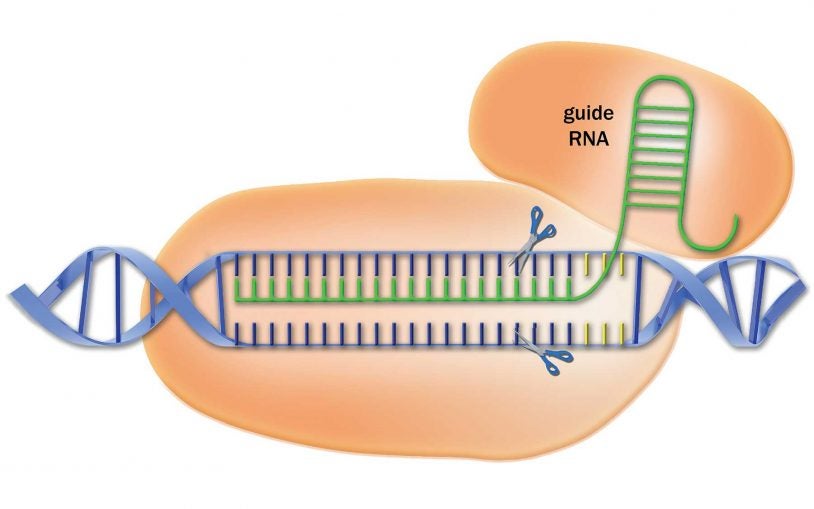
Library of CRISPR targeting sequences increases power and accuracy of the gene-editing method
July 20, 2017
New library of RNA sequences that can be used by researchers to direct the CRISPR-cas9 complex to cut DNA with unprecedented precision.
Newly identified small RNA fragments defend the genome when it’s ‘naked’
June 29, 2017
Fragments snipped from tRNAs protect embryonic stem cells while they’re being epigenetically reprogrammed.
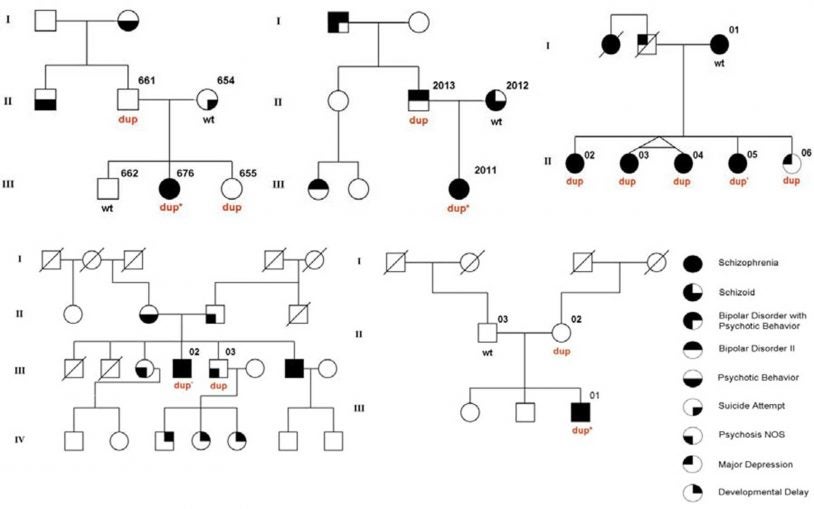
New statistical method finds shared ancestral gene variants involved in autism’s cause
June 21, 2017
Researchers find children with autism are genetically more like other autistic children than their unaffected siblings.
How cool is science? Students use barcoded DNA to identify a weapon against antibiotic resistance
June 14, 2017
Sometimes viruses cause harmful infections, but students in the DNA Learning Center’s Urban Barcode Research Program identified a new virus that could.
Detailed new ‘reference’ genome for maize shows the plant has deep resources for continued adaptation
June 12, 2017
A new, much more detailed reference genome for maize, or corn, as it is called in the U.S., provides new insight into the crop.
Base Pairs Episode 9.5: Redefining biologists, redefining genes
May 16, 2017
We talk with Molly Hammell about peering into deep space using a high-tech telescope, we also talk to Tom Gingeras about redefining the gene.
Base Pairs Episode 9: Dark matter of the genome, part 2
April 15, 2017
In this episode of Base Pairs, we question the mythos that is “junk DNA” and explore the mysterious non-coding portions of the genome.
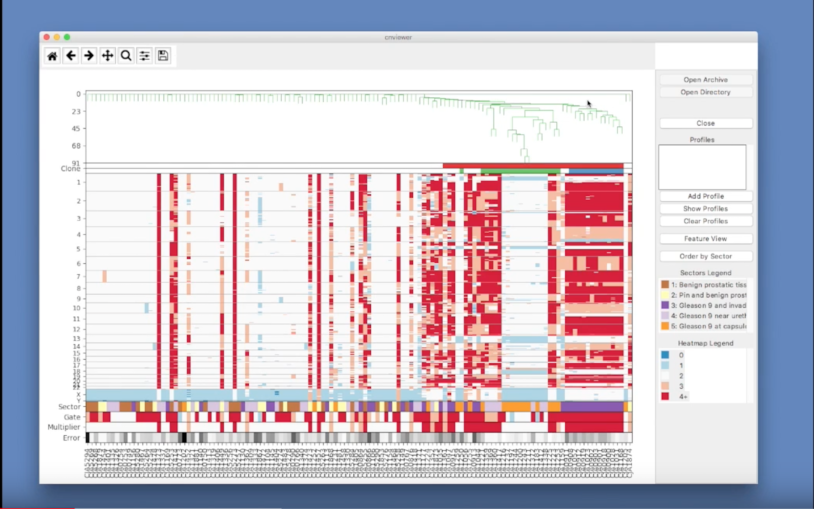
What is single-cell genome viewer?
March 31, 2017
Single-cell genome viewer is a graphical interface, integrating single-cell genomics and conventional pathology of tumors.
What a real-life science test looks like
March 24, 2017
By revealing evidence that contradicts the rationale for a new cancer drug, a pair of student scientists learns firsthand that when you do science.
Base Pairs Episode 8: Dark matter of the genome, part 1
March 15, 2017
In this episode, we explore how studying this unfamiliar territory could help scientists understand diseases such as ALS.
Dark matter of the genome, part 1
March 15, 2017
This episode of Base Pairs digs into "dark matter" a type of genetic information that could help scientists better understand diseases like
Quiescent cells brings to light the essential role of RNA interference
November 9, 2016
Professor Rob Martienssen presents evidence on the mechanisms that prompt cells to either to replicate their DNA or transcribe their DNA into RNA.
Focus on quiescent cells brings to light the essential role of RNA interference in transcription control
November 9, 2016
RNAi and other epigenetic processes induce changes in where and when specific genes are expressed without altering their genetic code.
How a cold-causing virus and inexplicable experiments helped revolutionize thinking about the genome
September 6, 2016
Nobel laureate Richard Roberts discusses the importance of RNA splicing and how scientists can learn from "failed" experiments.
Non-modern family
August 15, 2016
This episode on Base Pairs explores how genetic information to better understand human history.
Riding out of the shadows of ALS, toward better treatments
August 1, 2016
Graduate student Lisa Krug discusses her research and personal connection to ALS, and Ride for Life, a not-for-profit organization for ALS research.
To divide or not: A cellular feedback loop that enables new cells to make a fateful decision
July 21, 2016
Team explains a key mechanism to prevent aberrant cell proliferation
“Amazing protein diversity” is discovered in the maize plant
June 24, 2016
The genome of the corn plant “is a lot more exciting” than scientists have previously believed.
Unusual drug target generates exciting results in mouse models of metastatic breast cancer
May 17, 2016
In preliminary animal tests, targeting a RNA known as "Malat1" strikes an impressive blow against metastatic breast cancer.
Why is autism more common in boys?
April 29, 2016
Ivan Iossifov talks about some of the reasons why autism disproportionately effects boys on a genetic level.
What do autism “risk genes” do?
April 21, 2016
A discussion with Ivan Iossifov with what exactly the term "risk genes" means and what they do.
The CSHL Cancer Genomics group seeks to understand how the cancer genome differs from the normal genome, how these differences lead to the growth and development of cancer, and how biomarkers can be used for diagnosis and prognosis. Researchers in this area are members of the CSHL Cancer Center.
Much of the research focuses on the genomic changes that occur in numerous cancer types, such as breast, prostate, lung and pancreatic cancers, as well as leukemia, glioma and melanoma. Researchers have also focused on the development and application of sophisticated tools for genome analysis that permit high resolution mapping of deletions, amplifications and changes in the sequence or epigenetic status of chromosomal loci.
Another focus area is the development and application of genomic methods in cell culture and animal models. These include sophisticated chromosome engineering techniques as well as applications of large-scale RNAi and CRISPR screens to identify both driver genes and tumor-cell-specific dependencies, as well as single cell sequencing approaches. These genetic tools/approaches allow biological validation of loci discovered by analysis of cancer genomes.
A third area of focus centers on developing tools and software that can harness large scale genomic datasets that are available to the community. Examples include the study of allelic variation in the human genome and the development of computational methods for the discovery of cancer-associated genes and diagnostic cancer markers using genomic profiles derived from different tumor types. Many of these researchers are also part of the Simons Center for Quantitative Biology at CSHL.
The CSHL Plant Genomics group is using genomic approaches with the ultimate goal of improving access to food and fuel in the future. As part of Plant Biology research at CSHL, scientists are using genomic approaches to understand everything from plant evolution to how plants grow, develop, and reproduce.
This research is challenging because many plant genomes are very large. CSHL scientists have taken part in numerous plant genome sequencing projects including Arabidopsis, rice, sorghum and maize. In addition, CSHL plant scientists have participated in epigenomic sequencing and profiling. CSHL is also part of the iPlant Cyberinfrastructure consortium and the Long Island Biofuels Alliance.
Current research projects include sequencing the wheat genome through a combination of Illumina short read sequencing and long sequence reads using the new Pacific Biosciences sequencers. The latest results for this project can be found on the CSHL wheat genome sequencing project page.

Alexander Dobin
Next generation sequencing technologies revolutionized many areas of genetics and molecular biology, enabling quantitative analyses of the entire genomes and paving the way for Personalized Medicine. We develop novel statistical methods and computational algorithms for multi-omics processing and integration, and leverage Big Genomic Data to elucidate various problems in precision health, such as genetic and epigenetic mechanisms of cancer development and progression, and clinical impact of functional variants.

Thomas Gingeras
Only a small portion of the RNAs encoded in any genome are used to make proteins. My lab investigates what these noncoding RNAs (ncRNAs) do within and outside of cells, where regulators of their expression are located in the genome. This is particularly important in cancer. Our laboratory works on endometrial cancer and its relationship to age and obesity.

Sara Goodwin
I work on adapting and developing new methods/techniques for genome and transcriptome sequencing.

Christopher Hammell
As organisms develop, genes turn on and off with a precise order and timing, much like the order and duration of notes in a song. My group uses model organisms to understand the molecules that control the tempo of development. We also study how changes in the timing of gene expression contribute to diseases like cancer.

Ivan Iossifov
Every gene has a job to do, but genes rarely act alone. Biologists have built models of molecular interaction networks that represent the complex relationships between thousands of different genes. I am using computational approaches to help define these relationships, work that is helping us to understand the causes of common diseases including autism, bipolar disorder, and cancer.

David Jackson
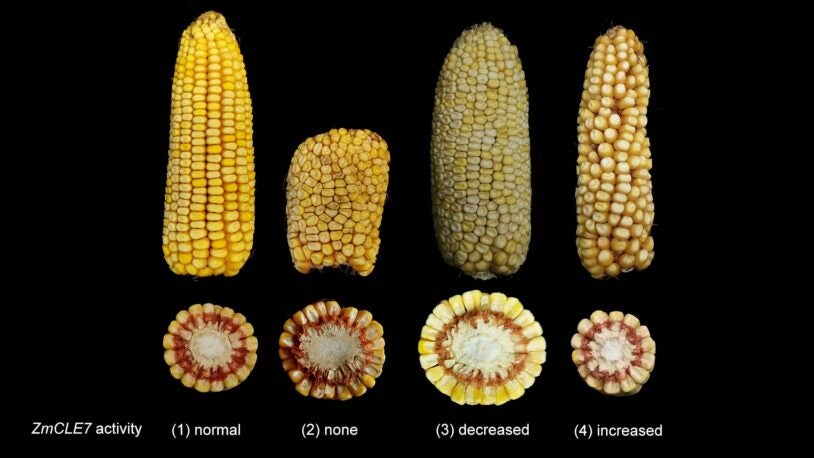
My lab studies genes and signals in cells that regulate the growth and shape of plants. We have discovered several genes that control plant architecture by exerting an influence on stem cells. By identifying the genes that control the number of stem cells in corn plants, for example, we’ve discovered a means of boosting the yield of that vital staple.

Alexander Krasnitz
Many types of cancer display bewildering intra-tumor heterogeneity on a cellular and molecular level, with aggressive malignant cell populations found alongside normal tissue and infiltrating immune cells. I am developing mathematical and statistical tools to disentangle tumor cell population structure, enabling an earlier and more accurate diagnosis of the disease and better-informed clinical decisions.

Dan Levy
We have recently come to appreciate that many unrelated diseases, such as autism, congenital heart disease and cancer, are derived from rare and unique mutations, many of which are not inherited but instead occur spontaneously. I am generating algorithms to analyze massive datasets comprising thousands of affected families to identify disease-causing mutations.

Zachary Lippman
My research team studies when and where, and how many branches, flowers, and fruits are produced on plants. All of plant development depends on small groups of stem cells at the tips of shoots known as meristems. By studying the genes that control stem cell production and maturation over space and time, within and between different developmental contexts, we are able to manipulate plant architecture and reproduction to improve crop yields.

Rob Martienssen
Chromosomes are covered with chemical modifications that help control gene expression. I study this secondary genetic code - the epigenome - and how it is guided by small mobile RNAs in plants and fission yeast. Our discoveries impact plant breeding and human health, and we use this and other genomic information to improve aquatic plants as a source of bioenergy.

W. Richard McCombie
Over the last two decades, revolutionary improvements in DNA sequencing technology have made it faster, more accurate, and much cheaper. We are now able to sequence up to 10 trillion DNA letters in just one month. I harness these technological advancements to assemble genomes for a variety of organisms and probe the genetic basis of neurological disorders, including autism and schizophrenia, better understand cancer progression and understand the complex structures of the genomes of higher plants.

Adam Siepel
I am a computer scientist who is fascinated by the challenge of making sense of vast quantities of genetic data. My research group focuses in particular on questions involving molecular evolution and transcriptional regulation, with applications to cancer and other diseases as well as to plant breeding and agriculture.

Zihua Wang
As a molecular biologist, I am fascinated by the new technologies in molecular genetic studies. My research is aimed at developing new technologies and applying them mainly to cancer studies, such as early detection, prognosis, and measuring minimal residual disease.

Doreen Ware
When we think of evolution, we often think about physical changes, like a plant developing broader leaves to collect more solar energy. Such evolution actually occurs within the plant’s DNA. I am using computational analysis and modeling to visualize how plant genomes have evolved over time, particularly those of staple crops. We are learning from this work to improve the range and yield of modern plants.

Michael Wigler
Devastating diseases like cancer and autism can be caused by spontaneous changes to our DNA—mutations first appearing in the child, or in our tissues as we age. We are developing methods to discover these changes in individuals, tumors, and even single cells, to promote early detection and treatments