Michael Wigler
Professor
Russell and Janet Doubleday Professor of Cancer Research
Cancer Center Member
Ph.D., Columbia University, 1978
wigler@cshl.edu | 516-367-8377
Devastating diseases like cancer and autism can be caused by spontaneous changes to our DNA—mutations first appearing in the child, or in our tissues as we age. We are developing methods to discover these changes in individuals, tumors, and even single cells, to promote early detection and treatments
Michael Wigler’s work provides a new paradigm for understanding and exploring human disease. The Wigler lab studies human cancer and the contribution of new mutation to genetic disorders. The cancer effort (with James Hicks, Alex Krasnitz, and Lloyd Trotman) focuses on breast and prostate cancers. It involves collaborative clinical studies to discover mutational patterns predicting treatment response and outcome and the development of diagnostics to detect cancer cells in bodily fluids such as blood and urine. The major tools are single-cell DNA and RNA analysis. The single-cell methods, which are in development, are also being applied to problems in neurobiology (with Josh Huang and Pavel Osten) to characterize neuronal subtypes, somatic mutation, and monoallelic expression. The Wigler lab’s genetic efforts are a collaboration with Ivan Iossifov and Dan Levy, and this team focuses on determining the role of new mutations in pediatric disorders. In a large-scale population sequencing project with W. Richard McCombie and the Genome Sequencing Center at Washington University in St. Louis, and supported by the Simons Foundation, the team has proven the contribution of this mechanism to autism. The work further suggests a relationship between the mutational targets in autism and the process of neuroplasticity that lies at the heart of learning. Smaller-scale population studies of congenital heart disease and pediatric cancer (collaborations with scientists at Columbia University and Memorial Sloan- Kettering Cancer Center, respectively) also point to new mutation as a causal factor in these disorders.
Empowering Insights: The science behind health
November 18, 2024
“The opportunity to turn curiosity into discoveries that impact the human condition is at the core of CSHL’s mission,” writes President Stillman.
Autism genetics: The faces behind the data
May 16, 2024
CSHL research on autism involves massive databases with thousands of genomes. Meet a few of the brave individuals who help make this work possible.
From cancer genetics to cancer treatments
January 18, 2024
One cancer gene, one cancer genome, two Cold Spring Harbor Laboratory discoveries that helped shape the face of modern cancer medicine.
The evolution of autism research
May 25, 2023
The conversation around autism has evolved over the past two decades. So has CSHL research. This retrospective shows how we’ve helped move the needle.
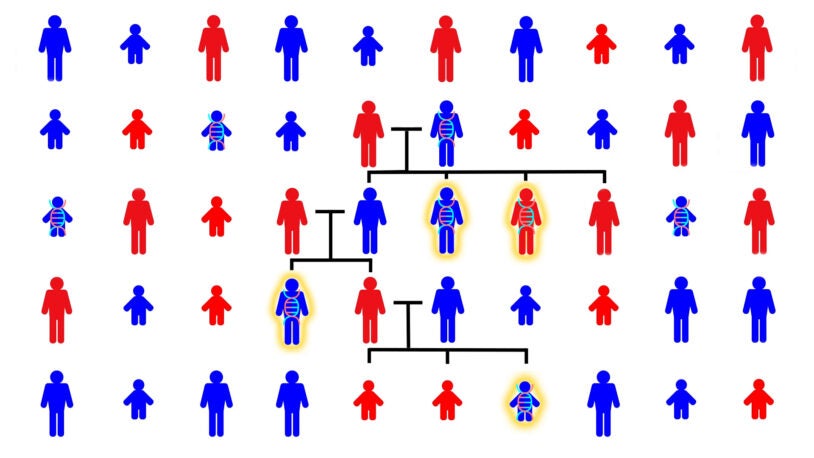
Autism in the family tree
May 23, 2023
CSHL scientists have studied the genetics of autism across hundreds of family trees. This animated video shows what they’ve found.
Siblings with autism share more of dad’s genome, not mom’s
May 22, 2023
CSHL study of more than 6,000 volunteer families overturns a long-held assumption about the genetic origins of autism spectrum disorder.
How well do you know autism spectrum disorders?
April 24, 2023
April is National Autism Awareness Month. Test your knowledge of autism spectrum disorders with this short quiz.
The genetic landscape of autism
August 16, 2022
CSHL Professor Michael Wigler discusses 20 years of research to paint a complete picture of the genetic causes of autism.
After 100 years of research, autism remains a puzzle
August 4, 2022
One geneticist is determined to piece together the causes of autism.
Tuveson and Wigler elected AACR Academy Fellows
May 12, 2020
CSHL Cancer Center Director David Tuveson and Professor Michael Wigler were chosen as 2020 Fellows of the AACR Academy.
Selected Publications
Integrated Computational Pipeline for Single-Cell Genomic Profiling
20 May 2020 | JCO clinical cancer informatics | 4:464-471
Chorbadjiev, L; Kendall, J; Alexander, J; Zhygulin, V; Song, J; Wigler, M; Krasnitz, A;
Novel Insights Into Breast Cancer Copy Number Genetic Heterogeneity Revealed by Single-Cell Genome Sequencing
13 May 2020 | eLife | 9:e51480
Baslan, T; Kendall, J; Volyanskyy, K; McNamara, K; Cox, H; D'Italia, S; Ambrosio, F; Riggs, M; Rodgers, L; Leotta, A; Song, J; Mao, Y; Wu, J; Shah, R; Gularte-Merida, R; Chadalavada, K; Nanjangud, G; Varadan, V; Gordon, A; Curtis, C; Krasnitz, A; Dimitrova, N; Harris, L; Wigler, M; Hicks, J;
Single-Chromosomal Gains Can Function as Metastasis Suppressors and Promoters in Colon Cancer
24 Feb 2020 | Developmental Cell | 52(4):413-428
Vasudevan, A; Baruah, P; Smith, J; Wang, Z; Sayles, N; Andrews, P; Kendall, J; Leu, J; Chunduri, N; Levy, D; Wigler, M; Storchova, Z; Sheltzer, J;
Multiplex Accurate Sensitive Quantitation (MASQ) With Application to Minimal Residual Disease in Acute Myeloid Leukemia
21 Feb 2020 | Nucleic Acids Research (NAR) | 48(7):e40
Moffitt, A; Spector, M; Andrews, P; Kendall, J; Alexander, J; Stepansky, A; Ma, B; Kolitz, J; Chiorazzi, N; Allen, S; Krasnitz, A; Wigler, M; Levy, D; Wang, Z;
Copolymerization of single-cell nucleic acids into balls of acrylamide gel
14 Nov 2019 | Genome Research | 30(1):49-61
Li, S; Kendall, J; Park, S; Wang, Z; Alexander, J; Moffitt, A; Ranade, N; Danyko, C; Gegenhuber, B; Fischer, S; Robinson, B; Lepor, H; Tollkuhn, J; Gillis, J; Brouzes, E; Krasnitz, A; Levy, D; Wigler, M;
All Publications
Dietary pro-oxidant therapy by a vitamin K precursor targets PI 3-kinase VPS34 function
25 Oct 2024 | Science | 386(6720):eadk9167
Swamynathan, Manojit; Kuang, Shan; Watrud, Kaitlin; Doherty, Mary; Gineste, Charlotte; Mathew, Grinu; Gong, Grace; Cox, Hilary; Cheng, Eileen; Reiss, David; Kendall, Jude; Ghosh, Diya; Reczek, Colleen; Zhao, Xiang; Herzka, Tali; Špokaitė, Saulė; Dessus, Antoine; Kim, Seung; Klingbeil, Olaf; Liu, Juan; Nowak, Dawid; Alsudani, Habeeb; Wee, Tse-Luen; Park, Youngkyu; Minicozzi, Francesca; Rivera, Keith; Almeida, Ana; Chang, Kenneth; Chakrabarty, Ram; Wilkinson, John; Gimotty, Phyllis; Diermeier, Sarah; Egeblad, Mikala; Vakoc, Christopher; Locasale, Jason; Chandel, Navdeep; Janowitz, Tobias; Hicks, James; Wigler, Michael; Pappin, Darryl; Williams, Roger; Cifani, Paolo; Tuveson, David; Laporte, Jocelyn; Trotman, Lloyd;
Sharing parental genomes by siblings concordant or discordant for autism
14 Jun 2023 | Cell Genomics | 3(6):100319
Wroten, Mathew; Yoon, Seungtai; Andrews, Peter; Yamrom, Boris; Ronemus, Michael; Buja, Andreas; Krieger, Abba; Levy, Dan; Ye, Kenny; Wigler, Michael; Iossifov, Ivan;
Magnetically Functionalized Hydrogels for High-Throughput Genomic Applications
1 Jan 2023 | Advanced Materials Technologies
Lammertse, E; Li, S; Kendall, J; Kim, C; Morris, P; Ranade, N; Levy, D; Wigler, M; Brouzes, E;
Accurate measurement of microsatellite length by disrupting its tandem repeat structure
12 Sep 2022 | Nucleic Acids Research (NAR) | :gkac723
Wang, Zihua; Moffitt, Andrea; Andrews, Peter; Wigler, Michael; Levy, Dan;
Targeted de novo phasing and long-range assembly by template mutagenesis
13 Jul 2022 | Nucleic Acids Research (NAR)
Li, Siran; Park, Sarah; Ye, Catherine; Danyko, Cassidy; Wroten, Matthew; Andrews, Peter; Wigler, Michael; Levy, Dan;