Ivan Iossifov
Professor
Ph.D., Columbia University, 2008
iossifov@cshl.edu | 516-367-6947
Faculty ProfileEvery gene has a job to do, but genes rarely act alone. Biologists have built models of molecular interaction networks that represent the complex relationships between thousands of different genes. I am using computational approaches to help define these relationships, work that is helping us to understand the causes of common diseases including autism, bipolar disorder, and cancer.
Ivan Iossifov focuses on the development of new methods and tools for genomic sequence analysis and for building and using molecular networks, and he applies them to specific biomedical problems. He studies the genetics of common diseases in humans using two main tools: next-generation sequencing and molecular networks representing functional relationships among genetic loci. These approaches in combination enable the kind of large-scale studies necessary for furthering our understanding of the complex etiology of disorders such as autism, bipolar disorder, and cancer.
Empowering Insights: The science behind health
November 18, 2024
“The opportunity to turn curiosity into discoveries that impact the human condition is at the core of CSHL’s mission,” writes President Stillman.
At the Lab Season 1 Research Rewind: Genetics
October 22, 2024
It’s the code for all life on Earth. This week At the Lab, we’re hacking it with the help of Cold Spring Harbor Laboratory’s geneticists.
Autism genetics: The faces behind the data
May 16, 2024
CSHL research on autism involves massive databases with thousands of genomes. Meet a few of the brave individuals who help make this work possible.
President’s essay: Bringing bold visions to life
May 26, 2023
CSHL President & CEO Bruce Stillman sees the Laboratory as a global hub for scientific expertise and a powerful launchpad for early-career scientists.
The evolution of autism research
May 25, 2023
The conversation around autism has evolved over the past two decades. So has CSHL research. This retrospective shows how we’ve helped move the needle.
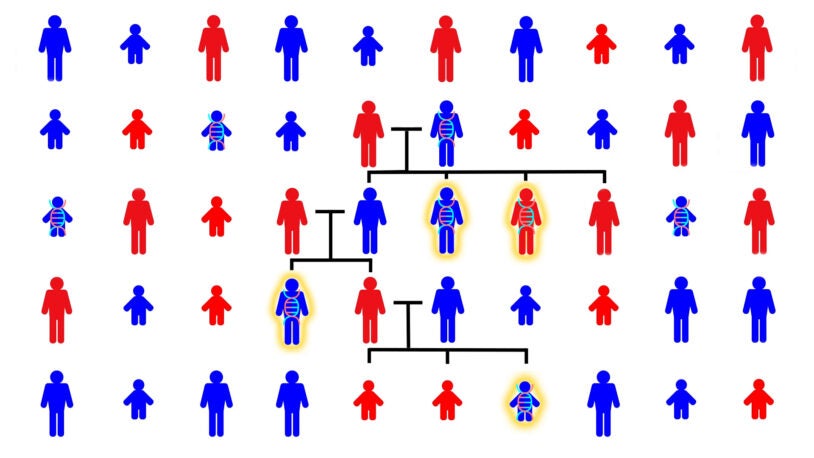
Autism in the family tree
May 23, 2023
CSHL scientists have studied the genetics of autism across hundreds of family trees. This animated video shows what they’ve found.
Siblings with autism share more of dad’s genome, not mom’s
May 22, 2023
CSHL study of more than 6,000 volunteer families overturns a long-held assumption about the genetic origins of autism spectrum disorder.
How well do you know autism spectrum disorders?
April 24, 2023
April is National Autism Awareness Month. Test your knowledge of autism spectrum disorders with this short quiz.
Portrait of a Neuroscience Powerhouse
April 27, 2018
A relatively small neuroscience group at CSHL is having an outsized impact on a dynamic and highly competitive field
Autism genetics study calls attention to impaired motor skills, general cognitive impairment
February 7, 2018
New research on the genetic causes of autism calls attention to diminished motor skills and suggests the importance of broad cognitive impairment
All Publications
The Genotype and Phenotypes in Families (GPF) platform manages the large and complex data at SFARI
11 Feb 2024 | bioRxiv
Chorbadjiev, Liubomir; Cokol, Murat; Weinstein, Zohar; Shi, Kevin; Fleisch, Chris; Dimitrov, Nikolay; Mladenov, Svetlin; Xu, Simon; Hall, Jake; Ford, Steven; Lee, Yoon-Ha; Yamrom, Boris; Marks, Steven; Munoz, Adriana; Lash, Alex; Volfovsky, Natalia; Iossifov, Ivan;
Heterozygous deletion of the autism-associated gene CHD8 impairs synaptic function through widespread changes in gene expression and chromatin compaction
5 Oct 2023 | American Journal of Human Genetics | 110(10):1750-1768
Shi, Xi; Lu, Congyi; Corman, Alba; Nikish, Alexandra; Zhou, Yang; Platt, Randy; Iossifov, Ivan; Zhang, Feng; Pan, Jen; Sanjana, Neville;
Genetic control of mRNA splicing as a potential mechanism for incomplete penetrance of rare coding variants
9 Aug 2023 | Genetics | 224(4):iyad115
Einson, Jonah; Glinos, Dafni; Boerwinkle, Eric; Castaldi, Peter; Darbar, Dawood; de Andrade, Mariza; Ellinor, Patrick; Fornage, Myriam; Gabriel, Stacey; Germer, Soren; Gibbs, Richard; Hersh, Craig; Johnsen, Jill; Kaplan, Robert; Konkle, Barbara; Kooperberg, Charles; Nassir, Rami; Loos, Ruth; Meyers, Deborah; Mitchell, Braxton; Psaty, Bruce; Vasan, Ramachandran; Rich, Stephen; Rienstra, Michael; Rotter, Jerome; Saferali, Aabida; Shoemaker, Moore; Silverman, Edwin; Smith, Albert; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; Mohammadi, Pejman; Castel, Stephane; Iossifov, Ivan; Lappalainen, Tuuli; Zhao, H;
Sharing parental genomes by siblings concordant or discordant for autism
14 Jun 2023 | Cell Genomics | 3(6):100319
Wroten, Mathew; Yoon, Seungtai; Andrews, Peter; Yamrom, Boris; Ronemus, Michael; Buja, Andreas; Krieger, Abba; Levy, Dan; Ye, Kenny; Wigler, Michael; Iossifov, Ivan;
Genetic control of mRNA splicing as a potential mechanism for incomplete penetrance of rare coding variants
31 Jan 2023 | bioRxiv
Einson, Jonah; Glinos, Dafni; Boerwinkle, Eric; Castaldi, Peter; Darbar, Dawood; de Andrade, Mariza; Ellinor, Patrick; Fornage, Myriam; Gabriel, Stacey; Germer, Soren; Gibbs, Richard; Hersh, Craig; Johnsen, Jill; Kaplan, Robert; Konkle, Barbara; Kooperberg, Charles; Nassir, Rami; Loos, Ruth; Meyers, Deborah; Mitchell, Braxton; Psaty, Bruce; Vasan, Ramachandran; Rich, Stephen; Rienstra, Michael; Rotter, Jerome; Saferali, Aabida; Shoemaker, M; Silverman, Edwin; Smith, Albert; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; Mohammadi, Pejman; Castel, Stephane; Iossifov, Ivan; Lappalainen, Tuuli;
Snakeobjects: an object-oriented workflow management system
12 Dec 2022 | bioRxiv
Yamrom, Boris; Lee, Yoon-ha; Marks, Steven; Chorbadjiev, Lubomir; Meyer, Hannah; Iossifov, Ivan;
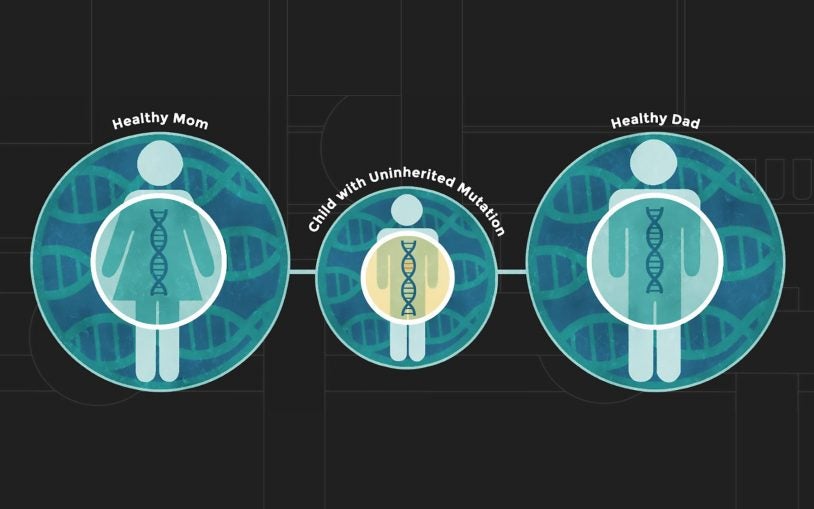
Rates of contributory de novo mutation in high and low-risk autism families.
1 Sep 2021 | Communications Biology | 4(1):1026
Yoon, Seungtai; Munoz, Adriana; Yamrom, Boris; Lee, Yoon-Ha; Andrews, Peter; Marks, Steven; Wang, Zihua; Reeves, Catherine; Winterkorn, Lara; Krieger, Abba; Buja, Andreas; Pradhan, Kith; Ronemus, Michael; Baldwin, Kristin; Levy, Dan; Wigler, Michael; Iossifov, Ivan;
Mapping and characterization of structural variation in 17,795 human genomes.
Jul 2020 | Nature | 583(7814):83-89
Abel, Haley; Larson, David; Regier, Allison; Chiang, Colby; Das, Indraniel; Kanchi, Krishna; Layer, Ryan; Neale, Benjamin; Salerno, William; Reeves, Catherine; Buyske, Steven; NHGRI Centers for Common Disease Genomics; Matise, Tara; Muzny, Donna; Zody, Michael; Lander, Eric; Dutcher, Susan; Stitziel, Nathan; Hall, Ira;
N-terminal Variant Asp14Asn of the Human p70 S6 Kinase 1 Enhances Translational Signaling Causing Different Effects in Developing and Mature Neuronal Cells
5 Mar 2020 | Neurobiology of Learning and Memory | 171(107203)
Venkatasubramani, J; Subramanyam, P; Pal, R; Reddy, B; Srinivasan, D; Chattarji, S; Iossifov, I; Klann, E; Bhattacharya, S;
De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms
24 Jan 2019 | European Journal of Human Genetics | 27(5):738-746
Jansen, S; van der Werf, I; Innes, A; Afenjar, A; Agrawal, P; Anderson, I; Atwal, P; van Binsbergen, E; van den Boogaard, M; Castiglia, L; Coban-Akdemir, Z; van Dijck, A; Doummar, D; van Eerde, A; van Essen, A; van Gassen, K; Guillen Sacoto, M; van Haelst, M; Iossifov, I; Jackson, J; Judd, E; Kaiwar, C; Keren, B; Klee, E; Klein Wassink-Ruiter, J; Meuwissen, M; Monaghan, K; de Munnik, S; Nava, C; Ockeloen, C; Pettinato, R; Racher, H; Rinne, T; Romano, C; Sanders, V; Schnur, R; Smeets, E; Stegmann, A; Stray-Pedersen, A; Sweetser, D; Terhal, P; Tveten, K; VanNoy, G; de Vries, P; Waxler, J; Willing, M; Pfundt, R; Veltman, J; Kooy, R; Vissers, Lelm; de Vries, B;