Alexander Krasnitz
Research Professor
Cancer Center Member
Ph.D., Tel Aviv University, 1990
krasnitz@cshl.edu | 516-367-6863
Faculty ProfileMany types of cancer display bewildering intra-tumor heterogeneity on a cellular and molecular level, with aggressive malignant cell populations found alongside normal tissue and infiltrating immune cells. I am developing mathematical and statistical tools to disentangle tumor cell population structure, enabling an earlier and more accurate diagnosis of the disease and better-informed clinical decisions.
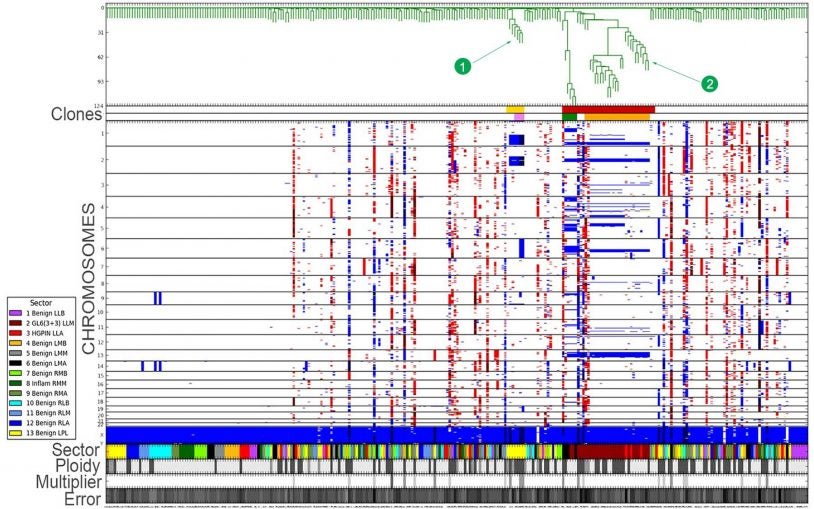
Alexander Krasnitz and colleagues develop mathematical and statistical tools to investigate population structure of cells comprising a malignant tumor and to reconstruct evolutionary processes leading up to that structure. These tools are designed to make optimal use of emerging molecular technologies, chief among them high-throughput genomic profiling of multiple individual cells harvested from a tumor. By analyzing these profiles, Krasnitz derives novel molecular measures of malignancy, such as the number of aggressive clones in a tumor, the invasive capacity of each clone and the amount of cancer-related genetic alteration sustained by clonal cells. Krasnitz and colleagues collaborate closely with clinical oncologists to explore the utility of such measures for earlier detection of cancer, more accurate patient outcome prediction and risk assessment, and better-informed choice of treatment options.
How evolved is your knowledge?
January 26, 2023
Test your knowledge of evolution with this quiz, inspired by the March 2023 performances of Isabella Rossellini’s play, Darwin’s Smile, at CSHL.
Unlocking cancer’s ancestry
December 27, 2022
New software may help reveal the complete connections between ancestry and cancer, which could lead to better, more personalized treatments.
New genetic research to understand racial disparity in cancers
September 8, 2020
Cold Spring Harbor Laboratory will study the genetic contributions of ethnicity to colon, endometrial, and pancreas cancers in African Americans.
New method to determine before surgery which prostate tumors pose a lethal threat
December 1, 2017
The news about prostate cancer can be confusing. It’s the third most common cancer type among Americans.
Next-gen cancer test
November 24, 2017
Knowing that cancers become lethal when they spread, investigators at Cold Spring Harbor Laboratory seek a way of detecting tumors much earlier
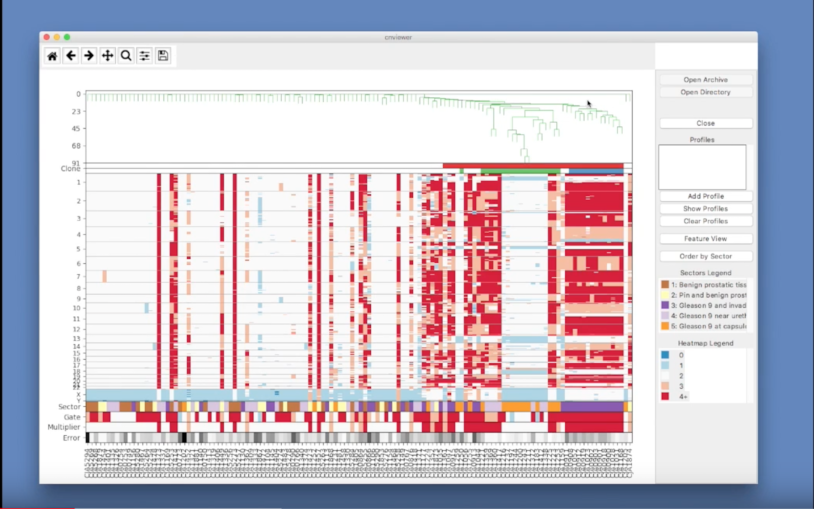
What is single-cell genome viewer?
March 31, 2017
Single-cell genome viewer is a graphical interface, integrating single-cell genomics and conventional pathology of tumors.
Breaking down breast cancer at CSHL
October 30, 2015
A look at how several researchers at CSHL contribute to the field of breast cancer research.
CSHL receives $50 million to establish Simons Center for Quantitative Biology
July 7, 2014
CSHL announced a $50 million gift from Jim and Marilyn Simons to establish the Simons Center for Quantitative Biology.
Mathematical technique de-clutters cancer-cell data, revealing tumor evolution, treatment leads
June 5, 2013
New technique reduces the burden of interpretation by identifying “cores of recurrent events.”
With new method, CSHL team is able to infer how tumors evolve and spread
March 11, 2011
Researchers find that cancerous tumors evolve in short, sudden bursts, rather than gradually.
All Publications
Correction: Genetic Ancestry Inference from Cancer-Derived Molecular Data across Genomic and Transcriptomic Platforms.
18 Jan 2023 | Cancer Research | 83(2):347
Belleau, Pascal; Deschênes, Astrid; Chambwe, Nyasha; Tuveson, David; Krasnitz, Alexander;
Genetic ancestry inference from cancer-derived molecular data across genomic and transcriptomic platforms
13 Dec 2022 | Cancer Research | :CAN-22
Belleau, Pascal; Deschênes, Astrid; Chambwe, Nyasha; Tuveson, David; Krasnitz, Alexander;
Ordered and deterministic cancer genome evolution after p53 loss
17 Aug 2022 | Nature
Baslan, Timour; Morris, John; Zhao, Zhen; Reyes, Jose; Ho, Yu-Jui; Tsanov, Kaloyan; Bermeo, Jonathan; Tian, Sha; Zhang, Sean; Askan, Gokce; Yavas, Aslihan; Lecomte, Nicolas; Erakky, Amanda; Varghese, Anna; Zhang, Amy; Kendall, Jude; Ghiban, Elena; Chorbadjiev, Lubomir; Wu, Jie; Dimitrova, Nevenka; Chadalavada, Kalyani; Nanjangud, Gouri; Bandlamudi, Chaitanya; Gong, Yixiao; Donoghue, Mark; Socci, Nicholas; Krasnitz, Alex; Notta, Faiyaz; Leach, Steve; Iacobuzio-Donahue, Christine; Lowe, Scott;
Accurate and robust inference of genetic ancestry from cancer-derived molecular data across genomic platforms
4 Feb 2022 | bioRxiv
Belleau, Pascal; Deschênes, Astrid; Tuveson, David; Krasnitz, Alexander;
Intraductal Transplantation Models of Human Pancreatic Ductal Adenocarcinoma Reveal Progressive Transition of Molecular Subtypes.
1 Oct 2020 | Cancer Discovery | 10(10):1566-1589
Miyabayashi, Koji; Baker, Lindsey; Deschênes, Astrid; Traub, Benno; Caligiuri, Giuseppina; Plenker, Dennis; Alagesan, Brinda; Belleau, Pascal; Li, Siran; Kendall, Jude; Jang, Gun; Kawaguchi, Risa; Somerville, Tim; Tiriac, Hervé; Hwang, Chang-Il; Burkhart, Richard; Roberts, Nicholas; Wood, Laura; Hruban, Ralph; Gillis, Jesse; Krasnitz, Alexander; Vakoc, Christopher; Wigler, Michael; Notta, Faiyaz; Gallinger, Steven; Park, Youngkyu; Tuveson, David;
SOAT1 promotes mevalonate pathway dependency in pancreatic cancer
7 Jul 2020 | Journal of Experimental Medicine (JEM) | 217(9)
Oni, T; Biffi, G; Baker, L; Hao, Y; Tonelli, C; Somerville, T; Deschênes, A; Belleau, P; Hwang, C; Sánchez-Rivera, F; Cox, H; Brosnan, E; Doshi, A; Lumia, R; Khaledi, K; Park, Y; Trotman, L; Lowe, S; Krasnitz, A; Vakoc, C; Tuveson, D;
Integrated Computational Pipeline for Single-Cell Genomic Profiling
20 May 2020 | JCO clinical cancer informatics | 4:464-471
Chorbadjiev, L; Kendall, J; Alexander, J; Zhygulin, V; Song, J; Wigler, M; Krasnitz, A;
Novel Insights Into Breast Cancer Copy Number Genetic Heterogeneity Revealed by Single-Cell Genome Sequencing
13 May 2020 | eLife | 9:e51480
Baslan, T; Kendall, J; Volyanskyy, K; McNamara, K; Cox, H; D'Italia, S; Ambrosio, F; Riggs, M; Rodgers, L; Leotta, A; Song, J; Mao, Y; Wu, J; Shah, R; Gularte-Merida, R; Chadalavada, K; Nanjangud, G; Varadan, V; Gordon, A; Curtis, C; Krasnitz, A; Dimitrova, N; Harris, L; Wigler, M; Hicks, J;
Multiplex Accurate Sensitive Quantitation (MASQ) With Application to Minimal Residual Disease in Acute Myeloid Leukemia
21 Feb 2020 | Nucleic Acids Research (NAR) | 48(7):e40
Moffitt, A; Spector, M; Andrews, P; Kendall, J; Alexander, J; Stepansky, A; Ma, B; Kolitz, J; Chiorazzi, N; Allen, S; Krasnitz, A; Wigler, M; Levy, D; Wang, Z;
Copolymerization of single-cell nucleic acids into balls of acrylamide gel
14 Nov 2019 | Genome Research | 30(1):49-61
Li, S; Kendall, J; Park, S; Wang, Z; Alexander, J; Moffitt, A; Ranade, N; Danyko, C; Gegenhuber, B; Fischer, S; Robinson, B; Lepor, H; Tollkuhn, J; Gillis, J; Brouzes, E; Krasnitz, A; Levy, D; Wigler, M;