My lab studies genes and signals in cells that regulate the growth and shape of plants. We have discovered several genes that control plant architecture by exerting an influence on stem cells. By identifying the genes that control the number of stem cells in corn plants, for example, we’ve discovered a means of boosting the yield of that vital staple.
David Jackson and colleagues study genes and signals that regulate plant growth and architecture. They are investigating a unique way in which plant cells communicate, by transporting regulatory proteins via small channels called plasmodesmata. These channels, which direct the flow of nutrients and signals through growing tissues, are regulated during development. The team discovered a gene encoding a chaperonin, CCT8, that controls the transport of a transcription factor SHOOTMERISTEMLESS (STM) between cells in the plant stem cell niche, or meristem. STM is critical for stem cell maintenance, and studies of the CCT8 gene indicate that movement of STM between cells is required for this function. The lab also continues to identify other genes that control plant architecture through effects on stem cell maintenance and identity, and their work has implications for crop yields. Recent examples include discovery of a subunit of a heterotrimeric G protein that is conserved throughout animals and plants, and their studies indicate that this gene controls stem cell proliferation. They have found that in plants, the G protein interacts with a completely different class of receptors than in animals. Their discovery helps to explain how signaling from diverse receptors is achieved in plants. This year, they also demonstrated that weak mutations in one of the receptor proteins can enhance seed production in maize, which could lead to yield increases. Separately, the lab has characterized system-wide networks of gene expression, using “next-gen” profiling and chromatin immunoprecipitation methods that have revealed many new hypotheses in developmental networks controlling inflorescence development. They are also developing a collection of maize lines that can drive expression of any reporter or experimental gene in any tissue type—tools of great interest to maize researchers that are being made available to the broader scientific community, enabling experiments never before possible in crop plants.
CSHL high schoolers named top Regeneron Scholars
March 3, 2025
CSHL volunteer Ishana Chadha and Partners for the Future participant Keita Takahashi are among the top student scientists in the U.S.
No Two Plants
February 11, 2025
CSHL postdoc’s experimental short film honors the legacy of Barbara McClintock, a continuing source of inspiration for women and girls in science.
Barbara McClintock’s corn
June 16, 2024
You’ve heard about Barbara McClintock’s Nobel Prize-winning research on corn genetics, but what about the corn itself?
Cocktails & Chromosomes: Best shots of 2023
January 16, 2024
After a four-year hiatus, Cocktails & Chromosomes returned in 2023. Relive the past year’s best moments and see what’s in store for 2024.
Cocktails & Chromosomes gets corny
August 10, 2023
In the best way possible! CSHL Professor David Jackson talked about corn genetics. Plus, we gave away tickets to Tony Award-winning musical Shucked.
CSHL’s Cocktails & Chromosomes series returns this summer
June 22, 2023
The event, serving up stimulating science talks over delicious drinks, heads to Industry bar in Huntington, New York. First on tap for June 29 is AI.
Digging up the deep-rooted secrets of perennial corn
March 10, 2023
Perennials may hold the key to sustainable farming. CSHL scientists are decoding the genes that let these plants withstand the test of time.
How tweaking genes keeps corn and rice on your plate
May 18, 2022
CSHL scientists leveraged lessons from the corn genome to improve another global staple crop, rice.
Do you have the dirt on plant research?
March 31, 2022
New research is constantly sprouting. Take this quiz and test your plant knowledge.
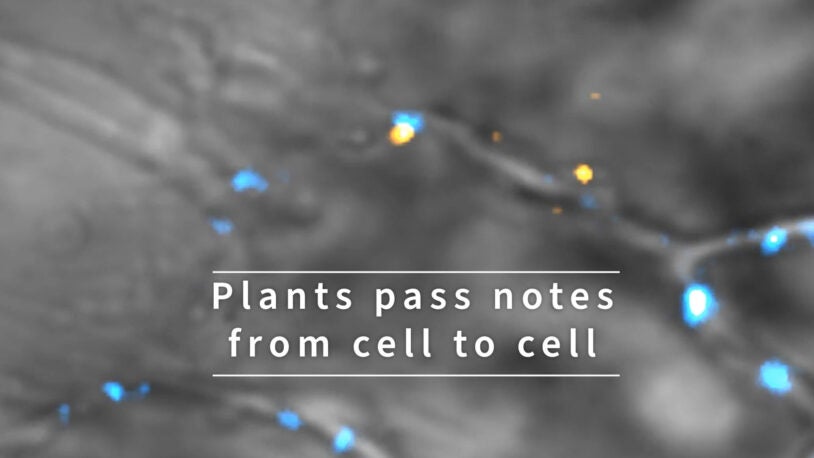
Psst! Plants pass notes from cell to cell
January 14, 2022
Plants relay messages critical for development between their cells using RNA. See tiny RNA messages zoom around inside plant cells.
All Publications
Maize2035: A decadal vision for intelligent maize breeding
17 Jan 2025 | Molecular Plant | :S1674-2052(25)00033
Liu, Hai-Jun; Liu, Jie; Zhai, Zhiwen; Dai, Mingqiu; Tian, Feng; Wu, Yongrui; Tang, Jihua; Lu, Yanli; Wang, Haiyang; Jackson, David; Yang, Xiaohong; Qin, Feng; Xu, Mingliang; Fernie, Alisdair; Zhang, Zuxin; Yan, Jianbing;
MaizeCODE reveals bi-directionally expressed enhancers that harbor molecular signatures of maize domestication
30 Dec 2024 | Nature Communications | 15(1):10854
Cahn, Jonathan; Regulski, Michael; Lynn, Jason; Ernst, Evan; de Santis Alves, Cristiane; Ramakrishnan, Srividya; Chougule, Kapeel; Wei, Sharon; Lu, Zhenyuan; Xu, Xiaosa; Ramu, Umamaheswari; Drenkow, Jorg; Kramer, Melissa; Seetharam, Arun; Hufford, Matthew; McCombie, W; Ware, Doreen; Jackson, David; Schatz, Michael; Gingeras, Thomas; Martienssen, Robert;
A Zea genus-specific micropeptide controls kernel dehydration in maize
7 Nov 2024 | Cell
Yu, Yanhui; Li, Wenqiang; Liu, Yuanfang; Liu, Yanjun; Zhang, Qinzhi; Ouyang, Yidan; Ding, Wenya; Xue, Yu; Zou, Yilin; Yan, Junjun; Jia, Anqiang; Yan, Jiali; Hao, Xinfei; Gou, Yujie; Zhai, Zhaowei; Liu, Longyu; Zheng, Yang; Zhang, Bao; Xu, Jieting; Yang, Ning; Xiao, Yingjie; Zhuo, Lin; Lai, Zhibing; Yin, Ping; Liu, Hai-Jun; Fernie, Alisdair; Jackson, David; Yan, Jianbing;
KRN5b regulates maize kernel row number through mediating phosphoinositol signalling
20 Sep 2024 | Plant Biotechnology Journal
Shen, Xiaomeng; Liu, Lei; Tran, Thu; Ning, Qiang; Li, Manfei; Huang, Liangliang; Zhao, Ran; Li, Yunfu; Qing, Xiangyu; Jackson, David; Bai, Yan; Song, Weibin; Lai, Jinsheng; Zhang, Zuxin; Zhao, Haiming;
Heterozygous fasciated ear mutations improve yield traits in inbred and hybrid maize lines
11 Sep 2024 | Plant Physiology | :kiae472
Wang, Jinbiao; Zheng, Qi; Zhang, Ruizhong; Huang, Zhaoyu; Wu, Qingyu; Liu, Lei; Ning, Qiang; Jackson, David; Xu, Fang;
Progressive meristem and single-cell transcriptomes reveal the regulatory mechanisms underlying maize inflorescence development and sex differentiation
13 Jun 2024 | Molecular Plant | 17(7):1019-1037
Sun, Yonghao; Dong, Liang; Kang, Lu; Zhong, Wanshun; Jackson, David; Yang, Fang;
The additive function of YIGE2 and YIGE1 in regulating maize ear length
28 May 2024 | The Plant Journal
Liu, Yu; Li, Huinan; Liu, Jie; Wang, Yuebin; Jiang, Chenglin; Zhou, Ziqi; Zhuo, Lin; Li, Wenqiang; Fernie, Alisdair; Jackson, David; Yan, Jianbing; Luo, Yun;
A spatial transcriptome map of the developing maize ear
14 May 2024 | Nature Plants
Wang, Yuebin; Luo, Yun; Guo, Xing; Li, Yunfu; Yan, Jiali; Shao, Wenwen; Wei, Wenjie; Wei, Xiaofeng; Yang, Tao; Chen, Jing; Chen, Lihua; Ding, Qian; Bai, Minji; Zhuo, Lin; Li, Li; Jackson, David; Zhang, Zuxin; Xu, Xun; Yan, Jianbing; Liu, Huan; Liu, Lei; Yang, Ning;
Tracking the messengers: Emerging advances in mRNA-based plant communication
24 Apr 2024 | Current Opinion in Plant Biology | 79:102541
Paul, Saikat; Jackson, David; Kitagawa, Munenori;
MaizeCODE reveals bi-directionally expressed enhancers that harbor molecular signatures of maize domestication
23 Feb 2024 | bioRxiv
Cahn, Jonathan; Regulski, Michael; Lynn, Jason; Ernst, Evan; Alves, Cristiane; Ramakrishnan, Srividya; Chougule, Kapeel; Wei, Sharon; Lu, Zhenyuan; Xu, Xiaosa; Drenkow, Jorg; Kramer, Melissa; Seetharam, Arun; Hufford, Matthew; McCombie, Richard; Ware, Doreen; Jackson, David; Schatz, Michael; Gingeras, Thomas; Martienssen, Robert;