Dan Levy
Associate Professor
Cancer Center Member
Ph.D., University of California, Berkeley 2005
levy@cshl.edu | 516-367-5039
Faculty ProfileWe have recently come to appreciate that many unrelated diseases, such as autism, congenital heart disease and cancer, are derived from rare and unique mutations, many of which are not inherited but instead occur spontaneously. I am generating algorithms to analyze massive datasets comprising thousands of affected families to identify disease-causing mutations.
There is increasing evidence that rare and unique mutations have a significant role in the etiology of many diseases such as autism, congenital heart disease, and cancer. Dan Levy’s group develops algorithms to identify these mutations from large, high-throughput data sets comprising thousands of nuclear families. After earlier working with high-resolution CGH arrays, Levy’s group now uses targeted sequence data. Levy has developed methods for identifying de novo mutations (i.e., those seen in a child but not in his or her parents) by simultaneously genotyping the entire family; the team is currently focused on building algorithms to detect copy-number variants and multiscale genomic rearrangements. Although their copy-number methods are based on “read” density, there are classes of mutations that require analysis at the level of the read. Thus, they are developing algorithms to identify insertions, deletions, inversions, transpositions, and other complex events. Other projects in the Levy lab include analysis of single-cell RNA, phylogenetic reconstruction from sparse data sets, and disentangling haplotypes from sperm and subgenomic sequence data.
Portrait of a Neuroscience Powerhouse
April 27, 2018
A relatively small neuroscience group at CSHL is having an outsized impact on a dynamic and highly competitive field
New study casts sharpest light yet on genetic mysteries of autism
October 29, 2014
Our picture of how genetic errors contribute to autism has just gotten sharper.
CSHL receives $50 million to establish Simons Center for Quantitative Biology
July 7, 2014
CSHL announced a $50 million gift from Jim and Marilyn Simons to establish the Simons Center for Quantitative Biology.
A striking link is found between the Fragile-X gene and mutations that cause autism
April 25, 2012
Scientists found a striking association between genes disrupted in children with autism and genes that are targets of FMRP
All Publications
Sharing parental genomes by siblings concordant or discordant for autism
14 Jun 2023 | Cell Genomics | 3(6):100319
Wroten, Mathew; Yoon, Seungtai; Andrews, Peter; Yamrom, Boris; Ronemus, Michael; Buja, Andreas; Krieger, Abba; Levy, Dan; Ye, Kenny; Wigler, Michael; Iossifov, Ivan;
Magnetically Functionalized Hydrogels for High-Throughput Genomic Applications
1 Jan 2023 | Advanced Materials Technologies
Lammertse, E; Li, S; Kendall, J; Kim, C; Morris, P; Ranade, N; Levy, D; Wigler, M; Brouzes, E;
Accurate measurement of microsatellite length by disrupting its tandem repeat structure
12 Sep 2022 | Nucleic Acids Research (NAR) | :gkac723
Wang, Zihua; Moffitt, Andrea; Andrews, Peter; Wigler, Michael; Levy, Dan;
Targeted de novo phasing and long-range assembly by template mutagenesis
13 Jul 2022 | Nucleic Acids Research (NAR)
Li, Siran; Park, Sarah; Ye, Catherine; Danyko, Cassidy; Wroten, Matthew; Andrews, Peter; Wigler, Michael; Levy, Dan;
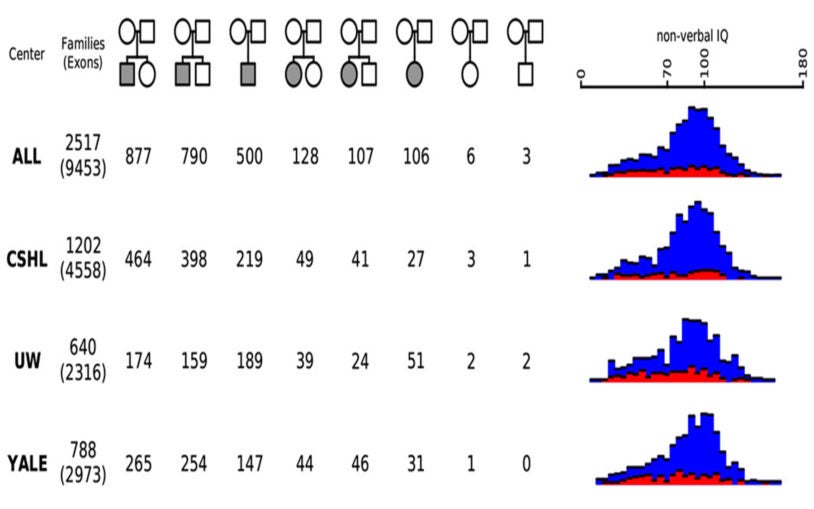
Rates of contributory de novo mutation in high and low-risk autism families.
1 Sep 2021 | Communications Biology | 4(1):1026
Yoon, Seungtai; Munoz, Adriana; Yamrom, Boris; Lee, Yoon-Ha; Andrews, Peter; Marks, Steven; Wang, Zihua; Reeves, Catherine; Winterkorn, Lara; Krieger, Abba; Buja, Andreas; Pradhan, Kith; Ronemus, Michael; Baldwin, Kristin; Levy, Dan; Wigler, Michael; Iossifov, Ivan;
Single-Chromosomal Gains Can Function as Metastasis Suppressors and Promoters in Colon Cancer
24 Feb 2020 | Developmental Cell | 52(4):413-428
Vasudevan, A; Baruah, P; Smith, J; Wang, Z; Sayles, N; Andrews, P; Kendall, J; Leu, J; Chunduri, N; Levy, D; Wigler, M; Storchova, Z; Sheltzer, J;
Multiplex Accurate Sensitive Quantitation (MASQ) With Application to Minimal Residual Disease in Acute Myeloid Leukemia
21 Feb 2020 | Nucleic Acids Research (NAR) | 48(7):e40
Moffitt, A; Spector, M; Andrews, P; Kendall, J; Alexander, J; Stepansky, A; Ma, B; Kolitz, J; Chiorazzi, N; Allen, S; Krasnitz, A; Wigler, M; Levy, D; Wang, Z;
Copolymerization of single-cell nucleic acids into balls of acrylamide gel
14 Nov 2019 | Genome Research | 30(1):49-61
Li, S; Kendall, J; Park, S; Wang, Z; Alexander, J; Moffitt, A; Ranade, N; Danyko, C; Gegenhuber, B; Fischer, S; Robinson, B; Lepor, H; Tollkuhn, J; Gillis, J; Brouzes, E; Krasnitz, A; Levy, D; Wigler, M;
Partial bisulfite conversion for unique template sequencing
25 Jan 2018 | Nucleic Acids Research (NAR) | 46(2):e10
Kumar, V; Rosenbaum, J; Wang, Z; Forcier, T; Ronemus, M; Wigler, M; Levy, D;
Utility of single cell genomics in diagnostic evaluation of prostate cancer
15 Jan 2018 | Cancer Research | 78(2):348-358
Alexander, J; Kendall, J; McIndoo, J; Rodgers, L; Aboukhalil, R; Levy, D; Stepansky, A; Sun, G; Chobadjiev, L; Riggs, M; Cox, H; Hakker, I; Nowak, D; Laze, J; Llukani, E; Srivastava, A; Gruschow, S; Yadav, S; Robinson, B; Atwal, G; Trotman, L; Lepor, H; Hicks, J; Wigler, M; Krasnitz, A;