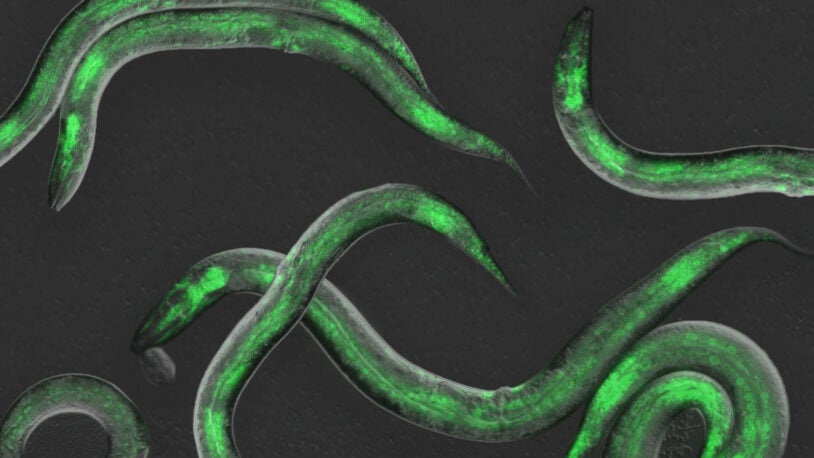
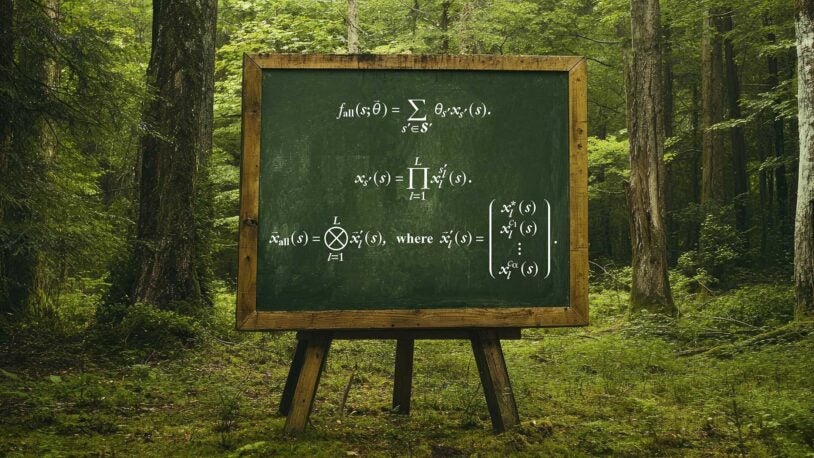Gene Regulation and Inheritance
 The Gene Regulation and Inheritance (GRI) Program investigates how DNA- and RNA-templated regulatory processes drive cancer development and lineage plasticity. The three main research themes of the GRI Program include: (1) defining cancer-specific roles of non-coding RNAs and RNA splicing; (2) revealing how chromatin regulation and genome inheritance support cancer development and maintenance; and (3) investigating how transcription factor master regulators drive cancer-specific gene programs.
The Gene Regulation and Inheritance (GRI) Program investigates how DNA- and RNA-templated regulatory processes drive cancer development and lineage plasticity. The three main research themes of the GRI Program include: (1) defining cancer-specific roles of non-coding RNAs and RNA splicing; (2) revealing how chromatin regulation and genome inheritance support cancer development and maintenance; and (3) investigating how transcription factor master regulators drive cancer-specific gene programs.
Program Co-leaders
Adrian Krainer, Ph.D.
Justin Kinney, Ph.D.
![]() The Gene Regulation and Inheritance Program combines experimental approaches with artificial intelligence (AI) research to uncover molecular mechanisms that control gene regulation and chromosomal inheritance. Program members are applying this knowledge toward innovative cancer therapeutic development. Areas of interest include uncovering mechanisms of splicing and RNA processing for developing antisense oligonucleotides as potential anti-cancer therapies, investigating chromatin-based regulation through histone modifications and nucleosome remodeling, and examining transcription factors through structural and mechanistic approaches. An expanding focus involves using genomic AI methods to decode how transcriptional control mechanisms are hijacked in cancer cells.
The Gene Regulation and Inheritance Program combines experimental approaches with artificial intelligence (AI) research to uncover molecular mechanisms that control gene regulation and chromosomal inheritance. Program members are applying this knowledge toward innovative cancer therapeutic development. Areas of interest include uncovering mechanisms of splicing and RNA processing for developing antisense oligonucleotides as potential anti-cancer therapies, investigating chromatin-based regulation through histone modifications and nucleosome remodeling, and examining transcription factors through structural and mechanistic approaches. An expanding focus involves using genomic AI methods to decode how transcriptional control mechanisms are hijacked in cancer cells.
Major achievements include developing antisense oligonucleotides for diffuse intrinsic pontine glioma, identifying BRD8 as a druggable target in glioblastoma, and creating organoid platforms to study triple negative breast cancer. The Program has also developed genomic AI methods for deciphering transcriptional regulation and elucidated how chromatin remodeling complexes reprogram DNA methylation. The ten Cancer Center Shared Resources are essential for this research, notably the Animal, Flow Cytometry, Microscopy, Mass Spectrometry, Functional Genomics, and Sequencing Technologies & Analysis Shared Resources.
School of Biological Sciences launches BioAI Ph.D. program
December 4, 2025
The new degree program will equip math, physics, and engineering students with the tools needed to make groundbreaking biomedical discoveries.
Shapeshifting cancers’ masters, unmasked
November 24, 2025
New research from CSHL’s Vakoc lab helps explain the bizarre behavior of certain pancreatic cancers and lung cancers that can change identities.
Joshua-Tor joins American Academy of Sciences and Letters
November 13, 2025
The prestigious group counts among its ranks Sir Salman Rushdie and three Nobel laureates, all with a background in chemistry.
Uplands Farm: Grounds for discovery
November 12, 2025
How a 12-acre research station is inspiring innovations needed to feed and fuel the world far into the future.
Breast cancer case study could inform clinical trials
November 10, 2025
CSHL researchers examined tissue samples from a 59-year-old woman diagnosed with stage 1 triple-negative breast cancer.
Dicer: Life’s ancient repair tool
October 28, 2025
In 2014, the Martienssen lab discovered that the Dicer protein helps protect the genome in humans and yeast. Now, they’ve figured out how.
Special edition: Breast Cancer Awareness Month
October 23, 2025
CSHL Professor David Spector discusses some of his lab’s latest research and the partnership making it possible.
A journey into animal aging
September 17, 2025
Time loops, babies with beards, and biology's favorite worm. CSHL Professor Christopher Hammel explores all these phenomena and more.
A graduation cap like no other
September 10, 2025
Group arts and crafts projects provide the Joshua-Tor lab a fun and memorable way to recognize its graduate students.
“Philifant” sculpture honors research leading to Spinraza
August 29, 2025
"Phil’s piece is more than a visual gift—it is a thoughtful expression of our mission to make life better through science,” says CSHL President Bruce Stillman
Inspiring breast cancer breakthroughs
August 18, 2025
For World Breast Cancer Research Day, see how CSHL scientists are leading the charge against this all-too-common disease.
A new fix for brain cancer?
August 14, 2025
Her research on glioblastoma was published in Nature and featured on ABC. Now, hear about her lab’s latest advances and high-impact collaborations.
NCI grants support ‘natural’ cancer research
August 11, 2025
CSHL’s Moses lab uses a technique called click chemistry to create new, nature-based compounds that could one day be used in the clinic.
Catalyst: A family legacy of hope
August 7, 2025
The Monti Foundation is supporting Dr. Lingbo Zhang's work in myelodysplastic syndrome (MDS) and has supported CSHL cancer research for nearly 20 years.
Can you ace this SMA quiz?
August 7, 2025
For Spinal Muscular Atrophy Awareness Month, test your knowledge of CSHL’s lifesaving SMA and RNA therapeutics research.
Giving hope to thousands of families
August 5, 2025
It’s SMA Awareness Month! See how CSHL’s fundamental biology research helped lead to the first successful treatment, and what's happened since.
Foundations for the Future: Topping-off ceremony
July 30, 2025
The CSHL community came together to put the final beam in place on a new Artificial Intelligence and Quantitative Biology building.
Licensed to live
July 29, 2025
How do we begin? Drawing from decades of research, CSHL President Bruce Stillman and colleagues offer a new lens to answer this fundamental question.
CSHL launches inaugural brain tumor meeting
July 17, 2025
The inaugural meeting brings together leading brain cancer researchers and clinicians to discuss the latest in the fight against these deadly diseases.
Adrian Krainer wins 2025 Heinrich Wieland Prize
July 16, 2025
Krainer is recognized for his lifesaving research on mRNA splicing and RNA therapeutics.
A new “link” to triple-negative breast cancer
June 30, 2025
There’s no effective treatment for the deadly disease. A discovery by CSHL Professor David Spector and grad student Wenbo Xu could point to the first.
The Adenovirus Gazebo
June 26, 2025
It’s not just any gazebo—it’s a meeting place, a scenic overlook, and a monument to CSHL’s decades of revolutionary genetics research.
Cocktails & Chromosomes: Heresy in genetics
June 17, 2025
Imagine everything you learned about DNA wasn’t quite wrong, but was only a fraction of the truth. Now, get more of the whole story.
A recipe to reverse cancer’s sweet tooth
June 16, 2025
Researchers reveal the first 3D structures of the kinase FN3K in different states, which may inform future therapeutics for a variety of cancers.
‘The bridge between hope and healing’
June 4, 2025
The Cosmopolitan Club hosts an inspiring event featuring CSHL Professor Alea Mills and Dr. John Boockvar of Netflix’s Lenox Hill and Emergency NYC.
President’s essay: We are science, and science is crucial
June 2, 2025
With public research funding under threat in the U.S., CSHL President Bruce Stillman outlines the stakes for people in our communities and abroad.
In nature’s math, freedoms are fundamental
May 28, 2025
CSHL quantitative biologists have developed a unified theory that could have countless applications, from plant breeding to drug discovery.
At the Lab: The human Microprocessor
May 27, 2025
There’s a Microprocessor inside you. Actually, there are trillions. Only they’re not computer processors. They’re much smaller and far more complex.
CSHL celebrates SBS Class of 2025
May 7, 2025
The CSHL School of Biological Sciences conferred nine Ph.D.s and one honorary Doctor of Science degree during its 22nd commencement ceremony.
The CSHL School of Biological Sciences’ class of 2025
May 5, 2025
The School of Biological Sciences awarded Ph.D. degrees to nine students this year. Read some of their stories and reflections on their time at CSHL.
Moses elected Fellow of Learned Society of Wales
April 29, 2025
The Welshman immigrated to the U.S. in 2020. He uses a revolutionary process called click chemistry to identify and test potential cancer treatments.
The food and fuel that farms itself
April 1, 2025
Duckweed can grow practically anywhere there’s sun and standing water. CSHL plant biologists identify the genes behind some of its most useful traits.
The natural evolution of bioRxiv and medRxiv
March 11, 2025
Independent nonprofit openRxiv launches to secure the long-term sustainability of biomedical research–sharing platforms bioRxiv and medRxiv.
CSHL receives $100K for leukemia research
February 24, 2025
The Don Monti Memorial Research Foundation’s donation supports the Zhang lab’s ongoing research on several variants of the deadly blood cancer.
Deciphering brain diseases with click chemistry
February 18, 2025
Cold Spring Harbor Laboratory Professors John Moses and Hiro Furukawa speak at the CSHL Association’s 2025 Annual Meeting.
Statement on NIH funding guidance
February 10, 2025
New NIH Guidance will severely reduce funding for research. Supporting science is an investment in our nation’s health, security, and economy.
Corn’s ancient ancestors are calling
February 6, 2025
Cold Spring Harbor Laboratory Professors Thomas Gingeras and Rob Martienssen have launched a new genomic encyclopedia called MaizeCODE.
Big clues emerge in small RNA mystery
December 16, 2024
How does the body separate “self” from “nonself?” CSHL Professor Rob Martienssen uncovers fascinating new leads in plant pollen and mouse sperm.
2024 Double Helix Medal recipient Dr. Katalin Karikó
December 10, 2024
She created the blueprint for mRNA vaccines. This year, Cold Spring Harbor Laboratory recognized her groundbreaking research and pioneering spirit.
The Microprocessor inside you
December 2, 2024
Stunning new images from CSHL’s Joshua-Tor lab reveal how the protein complex interacts with differently shaped primary microRNAs.
Frog Pond
November 27, 2024
These hopping amphibians aren’t the only animals that call the pond home. It’s a living snapshot of our area’s diverse ecosystem.
Empowering Insights: The science behind health
November 18, 2024
“The opportunity to turn curiosity into discoveries that impact the human condition is at the core of CSHL’s mission,” writes President Stillman.
The 2024 CSHL Raft Race
November 12, 2024
Ten teams brave cool harbor waters, the hot August sun, and new boat-building rules in the 9th annual CSHL Raft Race.
At the Lab Season 1 Research Rewind: AI+
October 29, 2024
This season’s final Research Rewind brings us from the realm of quantitative biology to neuroscience, genomics, and beyond.
At the Lab Season 1 Research Rewind: Genetics
October 22, 2024
It’s the code for all life on Earth. This week At the Lab, we’re hacking it with the help of Cold Spring Harbor Laboratory’s geneticists.
Adrian Krainer wins Albany Prize for biomedical research
October 8, 2024
A pioneer in the burgeoning field of RNA therapeutics, Krainer has now received America’s second-highest prize in medicine.
At the Lab Season 1 Research Rewind: Cancer
October 8, 2024
As the first season of our new podcast winds down, we’re revisiting all of our episodes with a focus on CSHL’s cutting-edge cancer research.
Plants have a backup plan
October 3, 2024
CSHL’s Rob Martienssen and his team discovered how plants like Arabidopsis continue to reproduce even when things go wrong in chromosome division.
At the Lab Episode 25: How maize became corn
September 24, 2024
Cold Spring Harbor Laboratory solves a plant biology mystery some 4,000 years in the making. The implications may go far beyond vegetables.
CSHL grad student wins International Birnstiel Award
September 23, 2024
Shushan Toneyan won the award for her thesis research in CSHL’s Koo lab. Toneyan is the co-creator of CREME, an AI-powered virtual laboratory.
At the Lab Episode 24: Putting the brakes on brain cancer
September 17, 2024
CSHL Professor Alea Mills compares the deadly brain cancer glioblastoma to a car with its brakes cut. Her lab works to reattach them.
Is CREME AI’s answer to CRISPR?
September 16, 2024
CREME, the latest AI toolkit from CSHL, is a virtual laboratory that may help scientists find new therapeutic targets in the genome.
At the Lab Episode 22: Outmuscling cancer
September 3, 2024
After 10 years, CSHL has made a breakthrough in the study of RMS, a rare pediatric cancer. How we got here is a story of innovation and perseverance.
Corn’s ‘missing link’
August 7, 2024
CSHL researchers have discovered a biological mechanism that may explain how corn spread so rapidly across the Americas 4,000 years ago.
At the Lab Episode 17: AI SQUID
July 30, 2024
Tune in to this week’s podcast to hear about the latest artificial intelligence model coming out of Cold Spring Harbor Laboratory.
At the Lab Episode 13: A more sustainable chemistry
July 2, 2024
For this week’s podcast, CSHL Professor John Moses bridges the gap between chemistry and biology in less than three minutes.
Table for 32
June 26, 2024
CSHL was excited to partner with Boy Scouts Troop 32 to help a young community member interested in science complete his Eagle Scout project.
SQUID pries open AI black box
June 21, 2024
CSHL’s Koo and Kinney labs have built a tool to suss out how AI analyzes the genome. What sets it apart? Decades of quantitative genetics knowledge.
Pancreatic cancer’s cellular amnesia
June 17, 2024
New study from CSHL Professor Christopher Vakoc and former postdoc Diogo Maia-Silva shows how basal-like cancer cells lose their original identity.
Barbara McClintock’s corn
June 16, 2024
You’ve heard about Barbara McClintock’s Nobel Prize-winning research on corn genetics, but what about the corn itself?
Wrexham University awards John Moses Honorary Fellowship
June 13, 2024
CSHL Professor John Moses returned to his hometown of Wrexham, Wales, where he was recognized for his contributions to science.
RNA splicing’s spotters
June 10, 2024
RNA therapeutics pioneer CSHL Professor Adrian Krainer has discovered a link between two important regulator proteins, SRSF1 and DDX23.
At the Lab Episode 10: The time of our lives
June 4, 2024
“You wouldn’t start making the fingernails on an arm until you had started to make the arm,” says CSHL’s Christopher Hammell. How’s that for a visual?
President’s essay: The continuous cycle of discovery
May 30, 2024
CSHL President & CEO Bruce Stillman discusses our institution’s societal impacts and global connections as forces for further scientific progress.
The CSHL School of Biological Sciences’ class of 2024
May 5, 2024
The School of Biological Sciences awarded Ph.D. degrees to 11 students this year. Here are some stories and reflections from their time at CSHL.
Hazen Tower
April 25, 2024
The Italian-style bell tower anchors CSHL’s Neuroscience Center. Its bell has rung out every hour on the hour, from 8 a.m. to 8 p.m., since 1991.
Nobel laureate honored at CSHL chemistry symposium
April 15, 2024
“The Future of Click Chemistry” brought together two-time Nobelist K. Barry Sharpless with his former apprentices John Moses and David Tuveson.
Up close and personal with cryo-EM
April 11, 2024
CSHL’s Cryo-Electron Microscopy course teaches the next generation of scientists to study life at the atomic level.
Click, click, boom—150 new molecules
April 4, 2024
CSHL Professor John Moses premieres an expansive line of new click chemistry products, uncovering leads for better antibiotics and cancer drugs.
CSHL’s Thomas Gingeras awarded $2 million NSF grant
April 3, 2024
Climate change threatens crops with acidic soils and aluminum toxicity. Gingeras leads an international team tackling this problem head-on.
From plant genomics to a bioscience revolution
March 25, 2024
CSHL played a lead role in mapping the first plant genome. Today, that breakthrough fuels a whole new understanding of life on Earth.
Cocktails & Chromosomes: Molecules to change the world
March 14, 2024
What is CSHL’s John Moses doing with that glowing liquid? Watch our expert chemist get a reaction from the crowd at Industry bar in Huntington, NY.
Why some RNA drugs work better than others
March 6, 2024
CSHL’s Justin Kinney and Spinraza inventor Adrian Krainer tested the newly approved SMA treatment, risdiplam, and another RNA therapeutic, branaplam.
Can AI uncover breast cancer risk factors?
February 26, 2024
This question lies at the heart of a new interdisciplinary collaboration between CSHL’s Camila dos Santos and Peter Koo.
How diet may impact cancer and possible treatments
February 1, 2024
Researchers at the CSHL Cancer Center study the links between disease and nutrition in hopes of uncovering new treatment and prevention strategies.
A quiz for the ages
January 29, 2024
Want to know the secret to a long life? So do CSHL scientists. Take this short quiz to see what they’ve found out about aging and longevity.
Joshua-Tor named CSHL Director of Research
January 2, 2024
The Cold Spring Harbor Laboratory professor and HHMI investigator steps into her new role effective January 2, 2024.
Dream big: A powerful vision for CSHL research
January 2, 2024
New Cold Spring Harbor Laboratory Director of Research Leemor Joshua-Tor shares her vision for the future of bioscience discovery.
CSHL celebrates 50th anniversary of McClintock Laboratory
November 20, 2023
Fifty years ago, CSHL honored Barbara McClintock by dedicating a building in her name. Today, it is home to four innovative cancer labs.
Breaking new ground: For science and society
November 13, 2023
CSHL’s Foundations for the Future campaign will propel the institution’s bioscience research and education programs to new heights and maximal impact.
Rob Martienssen awarded 2024 Genetics Society Medal
November 8, 2023
CSHL Professor Rob Martienssen earned the award for outstanding research contributions to the field of genetics.
First look: Whole-body gene expression
November 6, 2023
For the first time, scientists at CSHL have observed gene expression as it occurs throughout an animal. See life take shape in front of your eyes.
22nd Annual Women’s Partnership for Science Breaks Records
October 19, 2023
Cold Spring Harbor Laboratory held its 22nd Annual Women’s Partnership for Science lecture and luncheon.
Test your breast cancer awareness
October 18, 2023
Awareness is key to prevention and potential future treatments. Take this quiz to find out about the latest in breast cancer research at CSHL.
The dance of epigenetic inheritance
September 27, 2023
Making sure chromosomes get passed down correctly is hard work. Watch, through fluorescent and cryogenic lenses, how two proteins make it happen.
Stillman earns ASBMB Distinguished Scientist Award
September 7, 2023
The American Society for Biochemistry and Molecular Biology honored CSHL President & CEO Bruce Stillman for outstanding achievement in basic research.
These worms have rhythm
September 5, 2023
Observing gene expression in real time, CSHL scientists identified four molecules the C. elegans worm relies on to set the tempo of its development.
How plants pass down genetic memories
August 28, 2023
Thirty years ago, CSHL’s Rob Martienssen discovered plant gene DDM1. Now, he’s identified just how the DDM1 protein helps control inheritance.
The ancient viruses in human DNA
August 24, 2023
CSHL Assistant Professor Andrea Schorn takes us into her lab for a behind-the-scenes look at the parts of our genome that aren’t quite human.
Where did our DNA come from?
August 22, 2023
Half the human genome isn’t quite human. CSHL’s Andrea Schorn gives us the inside scoop on how our DNA turned ancient viruses into essential allies.
CSHL makes a splash with Swim Across America
August 21, 2023
Since 1987, the charity swim has raised over $100 million for cancer research. Here, CSHL Assistant Professor Semir Beyaz voices his support.
Blending history and science
August 17, 2023
One of the most famous experiments conducted at CSHL relied on the same state-of-the-art equipment found in kitchens across the U.S. in 1952—a Waring blender.
Laying the groundwork for drug discoveries
August 8, 2023
A new partnership between CSHL and one of the world’s leading biotech investors could streamline this process and help change society for the better.
Eight serving one: CSHL volleyball mid-season report
August 2, 2023
CSHL’s 32nd Volleyball League season sees eight teams battling for the coveted Tiernan Cup and a year’s worth of bragging rights.
Bite into this diet and disease quiz
July 5, 2023
Test your knowledge of how diet and nutrition affect health and disease with this short quiz.
Adrian Krainer awarded honorary IADR membership
June 21, 2023
Krainer was recognized for his pioneering research on spinal muscular atrophy and RNA therapeutics.
Lingbo Zhang wins National Institutes of Health MERIT Award
June 15, 2023
The highly prestigious award will support Zhang’s research on the role of nutrients and other environmental factors in blood cancer development.
CSHL harnesses biology’s favorite chemical
June 7, 2023
Life on Earth depends on phosphorus to give DNA structure. Soon, biology’s chosen chemical could make for new cancer treatments and green materials.
The digital dark matter clouding AI
June 5, 2023
Scientists have unknowingly encountered mysterious noise while using AI to decipher our genetic code. CSHL has found a way to cut through the fog.
Summer 2023 Harbor Transcript now online
May 31, 2023
This Special Annual Report edition of CSHL’s magazine provides a look back at some of the Laboratory’s biggest stories from 2022.
 Building publication list.
Building publication list.
Alexander Dobin
Next generation sequencing technologies revolutionized many areas of genetics and molecular biology, enabling quantitative analyses of the entire genomes and paving the way for Personalized Medicine. We develop novel statistical methods and computational algorithms for multi-omics processing and integration, and leverage Big Genomic Data to elucidate various problems in precision health, such as genetic and epigenetic mechanisms of cancer development and progression, and clinical impact of functional variants.

Thomas Gingeras
Only a small portion of the RNAs encoded in any genome are used to make proteins. My lab investigates what these noncoding RNAs (ncRNAs) do within and outside of cells, where regulators of their expression are located in the genome. This is particularly important in cancer. Our laboratory works on endometrial cancer and its relationship to age and obesity.

Christopher Hammell
As organisms develop, genes turn on and off with a precise order and timing, much like the order and duration of notes in a song. My group uses model organisms to understand the molecules that control the tempo of development. We also study how changes in the timing of gene expression contribute to diseases like cancer.

Leemor Joshua-Tor
Our cells depend on thousands of proteins and nucleic acids that function as tiny machines: molecules that build, fold, cut, destroy, and transport all of the molecules essential for life. My group is discovering how these molecular machines work, looking at interactions between individual atoms to understand how they activate gene expression, DNA replication, and small RNA biology.

Justin Kinney
Research in the Kinney Lab combines mathematical theory, machine learning, and experiments in an effort to illuminate how cells control their genes. These efforts are advancing the fundamental understanding of biology and biophysics, as well as accelerating the discovery of new treatments for cancer and other diseases.

Peter Koo
Deep learning has the potential to make a significant impact in basic biology and cancer, but a major challenge is understanding the reasons behind their predictions. My research develops methods to interpret this powerful class of black box models, with a goal of elucidating data-driven insights into the underlying mechanisms of sequence-function relationships.

Adrian R. Krainer
Our DNA carries the instructions to manufacture all the proteins needed by a cell. After each gene is copied from DNA into RNA, the RNA message is "spliced" - an editing process involving precise cutting and pasting. I am interested in how splicing normally works, how it is altered in genetic diseases and cancer, and how we can correct these defects for therapy.

Rob Martienssen
Chromosomes are covered with chemical modifications that help control gene expression. I study this secondary genetic code - the epigenome - and how it is guided by small mobile RNAs in plants and fission yeast. Our discoveries impact plant breeding and human health, and we use this and other genomic information to improve aquatic plants as a source of bioenergy.

David McCandlish
Some mutations are harmful but others are benign. How can we predict the effects of mutations, both singly and in combination? Using data from experiments that simultaneously measure the effects of thousands of mutations, I develop computational tools to predict the functional impact of mutations and apply these tools to problems in protein design, molecular evolution, and cancer.

Alea A. Mills
Cells employ stringent controls to ensure that genes are turned on and off at the correct time and place. Accurate gene expression relies on several levels of regulation, including how DNA and its associated molecules are packed together. I study the diseases arising from defects in these control systems, such as aging and cancer.

Lopa Mishra
My research focuses on the continuum of science-driven clinical care by working on novel therapies and improved clinical outcomes, honing liver disease, metabolism/alcohol, obesity/addiction gastrointestinal cancers, inflammatory bowel disease, and neural regulation of disease and cancer, which links to the field of bioelectronic medicine.

Andrea Schorn
Transposable elements make up half of our DNA. They control gene expression and have been a major evolutionary force in all organisms. The Schorn lab investigates how small RNAs identify and silence transposable elements when they become active during development and disease.

David L. Spector
The immense amount of DNA, RNA and proteins that contribute to our genetic programs are precisely organized inside the cell's nucleus. My group studies how nuclear organization impacts gene regulation, and how misregulation of non-coding RNAs contributes to human diseases such as cancer.

Bruce Stillman
Every time a cell divides, it must accurately copy its DNA. With 3 billion “letters” in the human genome, this is no small task. My studies reveal the many steps and molecular actors involved, as well as how errors in DNA replication are involved in diseases that range from cancer to rare genetic disorders.