David L. Spector
Professor
Robert B. Gardner Jr., Professor
Cancer Center Member
Ph.D., Rutgers University, 1980
spector@cshl.edu | 516-367-8456
The immense amount of DNA, RNA and proteins that contribute to our genetic programs are precisely organized inside the cell's nucleus. My group studies how nuclear organization impacts gene regulation, and how misregulation of non-coding RNAs contributes to human diseases such as cancer.
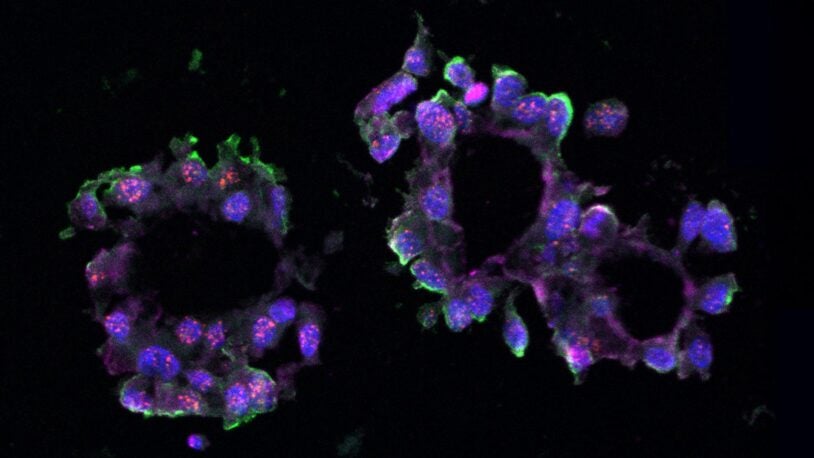
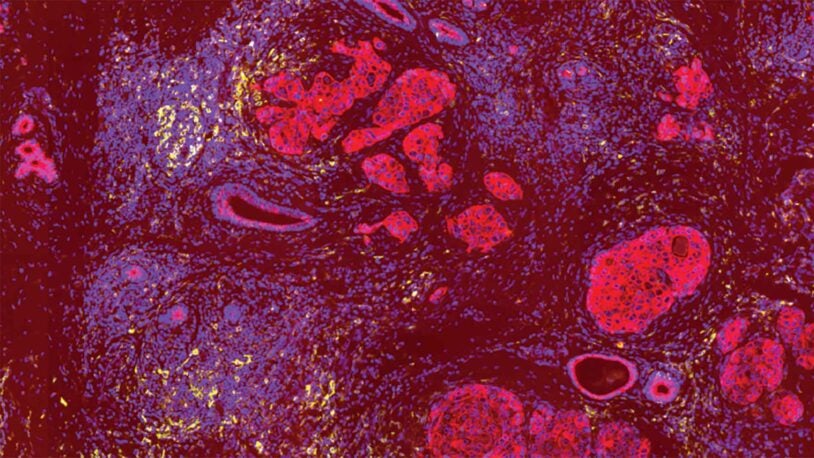
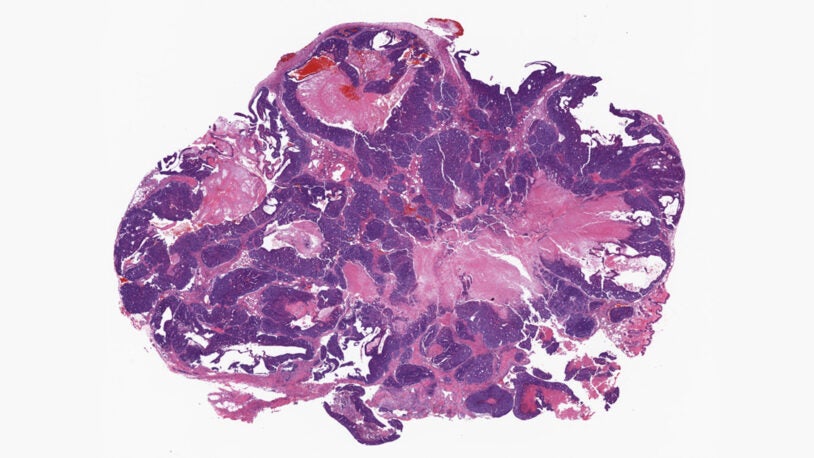
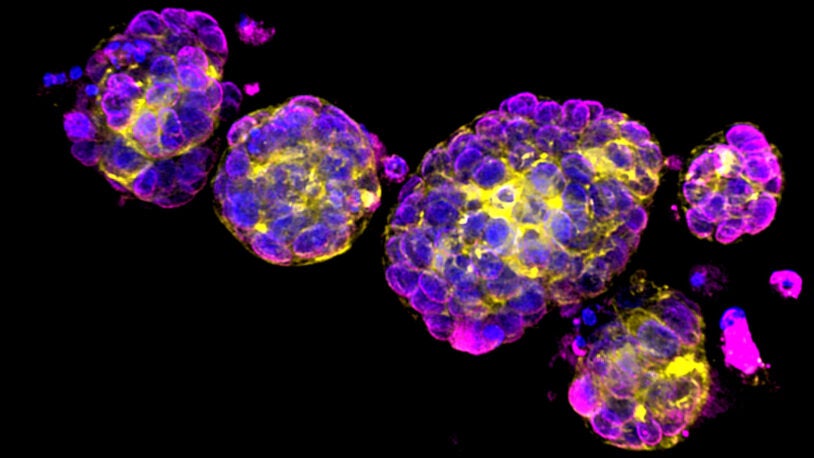
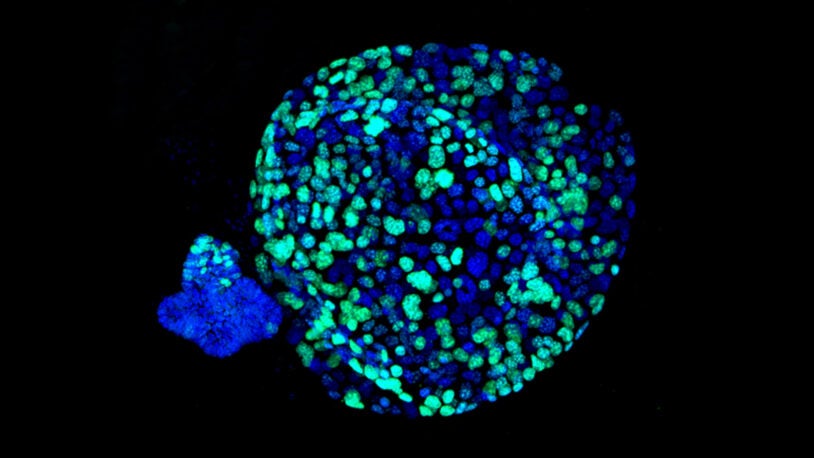
David L. Spector’s laboratory is focused on characterizing long non-coding RNAs (lncRNAs) that exhibit altered levels of expression in breast cancer progression and during embryonic stem cell differentiation. A major focus of their efforts has been on Malat1 lncRNA, which is one of the most abundant lncRNAs. The Spector lab previously identified a novel mechanism of 3′-end processing of this RNA. More recent studies have revealed that increased levels of Malat1 lncRNA impacts breast cancer progression and metastasis. Knockout or antisense oligonucleotide knockdown of Malat1 results in the differentiation of mammary tumors and a significant reduction in metastasis. Studies are currently under way to elucidate the mechanism of action of this abundant nuclear retained lncRNA and to implement innovative therapeutic approaches that can impact its function in vivo. In addition, they have developed and genomically and transcriptomically characterized patient-derived breast tumor organoid models from patients with triple negative breast cancer or invasive lobular carcinoma of the breast. These models are being used to identify and characterize additional lncRNAs that are over-expressed in breast cancer. Several candidates will be pursued at the functional level and as new therapeutic targets, as well as for drug development and screening in a patient-specific manner.
A second area of study in the Spector lab is focused on identifying and characterizing lncRNAs that play critical roles in pluripotency and/or lineage commitment, and to identify the specific pathways that they regulate. To do so they have reanalyzed the raw data from their previously published differential RNA-seq screen comparing lncRNA expression in naïve mouse embryonic stem cells (mESCs) vs neural progenitor cells (NPCs). They prioritized transcripts that were more than 2-fold upregulated in mESCs compared to NPCs. This resulted in the identification of 147 mESC specific transcripts of which 105 have potential human orthologs. From this analysis several exciting candidates have been prioritized and are being pursued at the functional level. These studies will provide new insights as to how regulatory signals instructed by lncRNAs can impact pluripotency and/or lineage commitment which will influence basic biology and/or disease states.
A new “link” to triple-negative breast cancer
June 30, 2025
There’s no effective treatment for the deadly disease. A discovery by CSHL Professor David Spector and grad student Wenbo Xu could point to the first.
Joshua-Tor named CSHL Director of Research
January 2, 2024
The Cold Spring Harbor Laboratory professor and HHMI investigator steps into her new role effective January 2, 2024.
Test your breast cancer awareness
October 18, 2023
Awareness is key to prevention and potential future treatments. Take this quiz to find out about the latest in breast cancer research at CSHL.
Cancer lab makes surprise discoveries in heart disease
November 30, 2022
Two separate studies from the Spector lab at CSHL suggest that certain genes can lead to cardiac problems.
Can cancer be treated by changing its cells?
April 11, 2022
Tumors grow when cells lose their biological identity. A promising therapeutic might restore their sense of self.
Paving a path to triple-negative breast cancer treatment
March 29, 2022
Researchers collected a biobank of triple-negative breast cancer mini-tissues to search for new and potentially patient-specific treatments.
Regulatory RNAs promote breast cancer metastasis
December 22, 2020
A gene-regulating bit of RNA promotes breast cancer metastasis. Agents that destroy that RNA provide hope for a new drug.
One experiment: Organoids as living laboratories
October 30, 2020
These tiny balls of cells are revolutionizing the research and treatment of pancreas and other types of cancers.
Women’s coalition donates $100k to breast cancer research
December 6, 2018
Members of the Manhasset Women’s Coalition Against Breast Cancer supported research through a donation from the Ladies Night Out fundraiser.
Undergrad symposium highlights lessons in the lab
August 20, 2018
The 2018 Undergraduate Research Program class presents their projects after working for ten weeks in various labs across campus.
Selected Publications
Platr4 is an early embryonic lncRNA that exerts its function downstream on cardiogenic mesodermal lineage commitment
7 Nov 2022 | Developmental Cell | 57(21):2450-2468.e7
Hazra, Rasmani; Brine, Lily; Garcia, Libia; Benz, Brian; Chirathivat, Napon; Shen, Michael; Wilkinson, John; Lyons, Scott; Spector, David;
Patient-derived triple-negative breast cancer organoids provide robust model systems that recapitulate tumor-intrinsic characteristics
18 Feb 2022 | Cancer Research
Bhatia, Sonam; Kramer, Melissa; Russo, Suzanne; Naik, Payal; Arun, Gayatri; Brophy, Kyle; Andrews, Peter; Fan, Cheng; Perou, Charles; Preall, Jonathan; Ha, Taehoon; Plenker, Dennis; Tuveson, David; Rishi, Arvind; Wilkinson, John; McCombie, W; Kostroff, Karen; Spector, David;
Mammary Tumor-Associated RNAs Impact Tumor Cell Proliferation, Invasion, and Migration
27 Sep 2016 | Cell Reports | 17(1):261-274
Diermeier, Sarah; Chang, Kung-Chi; Freier, Susan; Song, Junyan; El Demerdash, Osama; Krasnitz, Alexander; Rigo, Frank; Bennett, C; Spector, David;
The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult
2012 | Cell Reports | 2(1):111-123
Zhang, B; Arun, G; Mao, Y; Lazar, Z; Hung, G; Bhattacharjee, G; Xiao, X; Booth, C; Wu, J; Zhang, C; Spector, D;
All Publications
The unique catalytic properties of PSAT1 mediate metabolic adaptation to glutamine blockade
Aug 2024 | Nature Metabolism | 6(8):1529-1548
Qiu, Yijian; Stamatatos, Olivia; Hu, Qingting; Ruiter Swain, Jed; Russo, Suzanne; Sann, Ava; Costa, Ana; Violante, Sara; Spector, David; Cross, Justin; Lukey, Michael;
MARK2/MARK3 kinases are catalytic co-dependencies of YAP/TAZ in human cancer
26 Jul 2024 | Cancer Discovery
Klingbeil, Olaf; Skopelitis, Damianos; Tonelli, Claudia; Yoshimoto, Toyoki; Alpsoy, Aktan; Panepinto, Maria; Minicozzi, Francesca; Merrill, Joseph; Cafiero, Amanda; Aggarwal, Disha; Russo, Suzanne; Ha, Taehoon; Demerdash, Osama; Wee, Tse-Luen; Spector, David; Lyons, Scott; Tuveson, David; Cifani, Paolo; Vakoc, Christopher;
Interaction between MED12 and ΔNp63 activates basal identity in pancreatic ductal adenocarcinoma
17 Jun 2024 | Nature Genetics
Maia-Silva, Diogo; Cunniff, Patrick; Schier, Allison; Skopelitis, Damianos; Trousdell, Marygrace; Moresco, Philip; Gao, Yuan; Kechejian, Vahag; He, Xue-Yan; Sahin, Yunus; Wan, Ledong; Alpsoy, Aktan; Liverpool, Jynelle; Krainer, Adrian; Egeblad, Mikala; Spector, David; Fearon, Douglas; Dos Santos, Camila; Taatjes, Dylan; Vakoc, Christopher;
MARK2/MARK3 kinases are catalytic co-dependencies of YAP/TAZ in human cancer
28 Feb 2024 | bioRxiv
Klingbeil, Olaf; Skopelitis, Damianos; Tonelli, Claudia; Alpsoy, Aktan; Minicozzi, Francesca; Aggarwal, Disha; Russo, Suzanne; Ha, Taehoon; Demerdash, Osama; Spector, David; Tuveson, David; Cifani, Paolo; Vakoc, Christopher;
Chronic stress increases metastasis via neutrophil-mediated changes to the microenvironment
13 Feb 2024 | Cancer Cell
He, Xue-Yan; Gao, Yuan; Ng, David; Michalopoulou, Evdokia; George, Shanu; Adrover, Jose; Sun, Lijuan; Albrengues, Jean; Daßler-Plenker, Juliane; Han, Xiao; Wan, Ledong; Wu, Xiaoli; Shui, Longling; Huang, Yu-Han; Liu, Bodu; Su, Chang; Spector, David; Vakoc, Christopher; Van Aelst, Linda; Egeblad, Mikala;
A sub-set of guanine- and cytosine-rich genes are actively transcribed at the nuclear Lamin B1 region
28 Oct 2023 | bioRxiv
Balasooriya, Gayan; Wee, Tse-Luen; Spector, David;