Knowing that cancers become lethal when they spread, investigators at Cold Spring Harbor Laboratory (CSHL) seek a way of detecting tumors much earlier than now possible—when they’re more likely to be curable. Fleshing out an idea Professor Michael Wigler had years ago—before there was technology to act on it—research led by Associate Professor Alexander Krasnitz has advanced an inexpensive method to do just this. It begins with a simple blood draw like that performed at an annual physical.
X-rays, MRI and CT scans, as well as biopsies, have helped detect certain cancers earlier. The next-generation technology is a blood test—sensitive enough to consistently find the earliest, invisible cancers, and accurate enough to avoid false positives and negatives.
Publishing in Trends in Molecular Medicine this year, Krasnitz, Wigler and colleagues proposed such a blood screen. They tested it “in silico,” i.e., by plugging real data into a mathematical model they devised. The data came from 3,852 patients with 11 types of solid tumors, whose cases are archived in The Cancer Genome Atlas (TCGA). The results were encouraging, suggesting that the CSHL early detection method is feasible—both accurate and sensitive enough to find many kinds of solid cancers, perhaps when they are still invisible, and at around $1,000 per test—a number certain to come down with the cost of DNA sequencing.
There is a critical supposition in the team’s demonstration. Buoyed by a growing body of evidence that even very early tumors shed cells into the blood, they assume that 10 related tumor cells will be present in a typical 50 billion-cell 10 ml blood draw (about 2 teaspoons’ worth). The team assessed their proposed method by assuming it was able to find 6 of the 10 expected tumor cells in a known cancer patient’s blood sample.
The computerized test of the CSHL early detection method showed that it detects most common solid tumor types with great sensitivity. Krasnitz and colleagues calculated they would have detected nearly 100% of the 457 ovarian cancers and 261 small-cell lung cancers in the TCGA sample; roughly 90% of the 750 breast cancers, and 70%–75% of the 485 glioblastomas and 349 colon tumors. Tumors that tend to have fewer copy number variations, or CNVs—one of the two cancer hallmarks the method is based on—were harder to detect: about 55% of 373 kidney tumors and 378 uterine tumors. But the method’s ability to accurately detect very early ovarian and lung cancers is particularly good news, since both are notorious for being diagnosed at an advanced stage.
The CSHL method is distinguished from other “liquid biopsy” tests now in development, which sift for fragments of mutated DNA floating in the blood. The weakness of that approach, says the CSHL team, is precisely its dependence upon fragmentary evidence—“cell-free” DNA that has detached from the cells it came from. “It usually takes multiple genetic hits to initiate cancer,” Krasnitz explains, “and one problem with looking at cell-free DNA is that you don’t know, assuming you find two or more suspicious fragments, whether they came from the same cell or not. If they don’t, the ‘signal’ could be spurious.”
To be useful, Krasnitz stresses, clinicians need to be able to follow up on positive results in a meaningful way. Because it is based on the capture and analysis of intact cells, the CSHL method can readily trace suspect cells to their tissue of origin, by reading their epigenetic profile (chemical marks on the DNA that vary by cell type) or by sequencing RNAs generated by active genes (similarly varying according to cell and tissue type). These tests add cost, but would only be performed in the minority of cases in which there is evidence of clonality—a second cancer hallmark necessary for a positive result in the CSHL method [see “How it works,” below].
If closer analysis points to the lung, or to the breast, for example, still more expensive high-resolution scans and biopsies can be used to spot early tumors, which might then be treated and possibly cured. Unlike most biopsies now performed, those occasioned by the new screening method would likely be ordered before a single symptom appeared—as the result of a routine screen performed at an annual check-up.
This is precisely the grail that the medical community has long sought to find and treat cancers before they become lethal. The next step, says Krasnitz, is to test the accuracy of the method in a clinical study involving patients just diagnosed with cancer. The use of a blood screen as a cancer preventive given at a yearly physical is still a few years down the road. It would first be used in patients who have already been treated for cancer, to detect recurrence, or as a preventive in people known to be at high risk for particular cancer types, such as HER-2-positive breast cancer.
How it works
The CSHL early detection method features a technology called single-cell sequencing, which generates a genome-wide copy number profile of each cell. These profiles register copy number variations, or CNVs: places in the genome where there is extra or missing DNA. Everyone has CNVs in their genome, but they’re almost always harmless. In cancer cells, the genome is highly disturbed and CNVs abound. An irregular copy number profile is one of the hallmarks of cancer that the CSHL early detection method looks for.
The CSHL method also looks for clonality, a cancer hallmark that cannot be discerned by studying cell-free DNA. A clone is a group of genetically identical cells that share a common ancestor. From a single clonal population of aberrant cells, cancers advance in punctuated, staccato-like bursts. Cells that continue to mutate gain a survival advantage—one reason cancer is so hard to defeat.
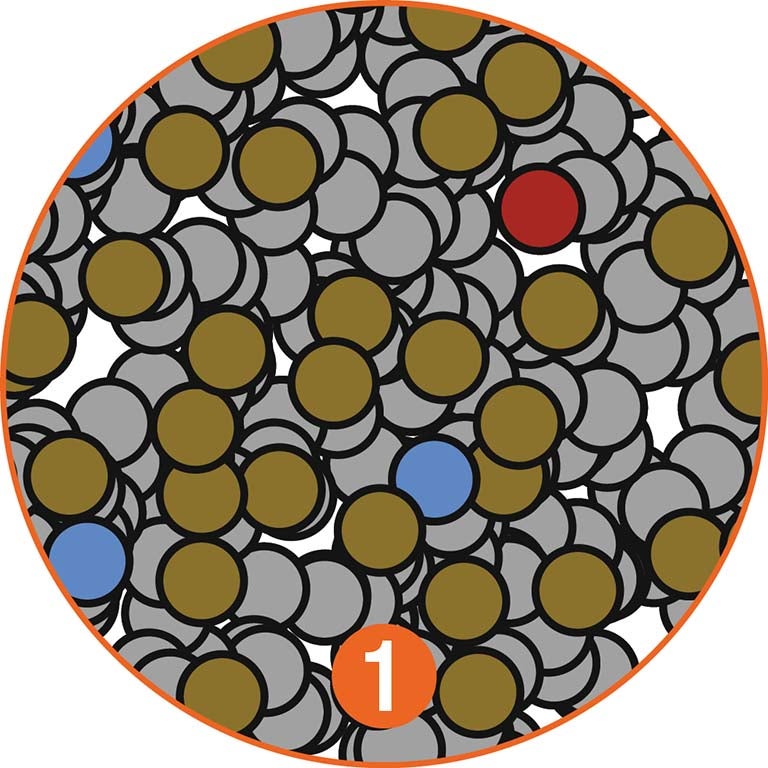
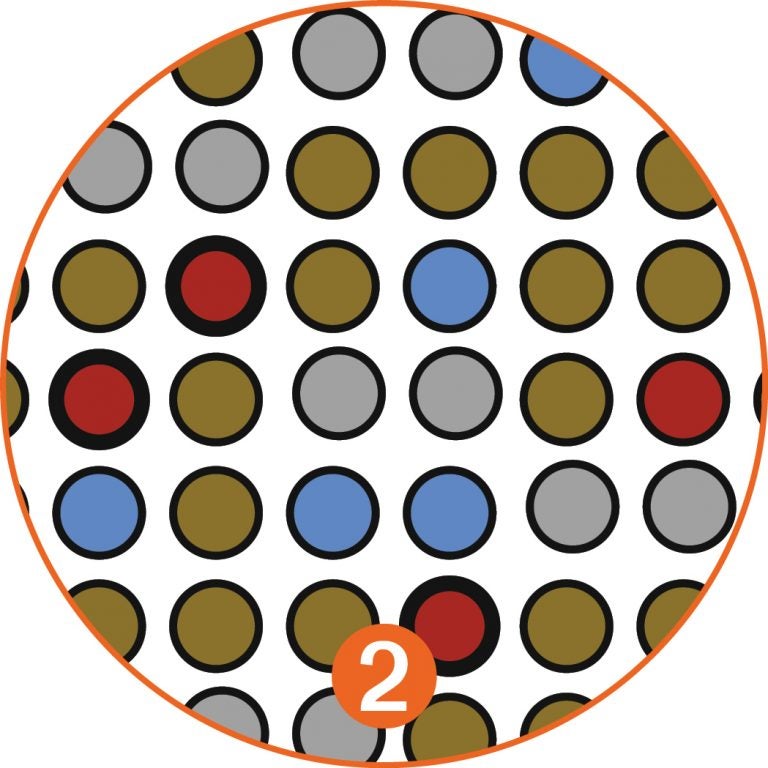
In the early detection method proposed by Krasnitz and Wigler, an initial screen reduces the 50 billion cells of the initial blood draw [1] to a much more manageable “enriched” population of a few thousand cells. It does this by filtering out nearly all of the blood cells [2]. Most non-cancerous white blood cells and tissue cells shed into the blood are eliminated in the second stage of the analysis, which looks at copy number variations, because the CNV profiles of these cells are normal: flat lines in [3].
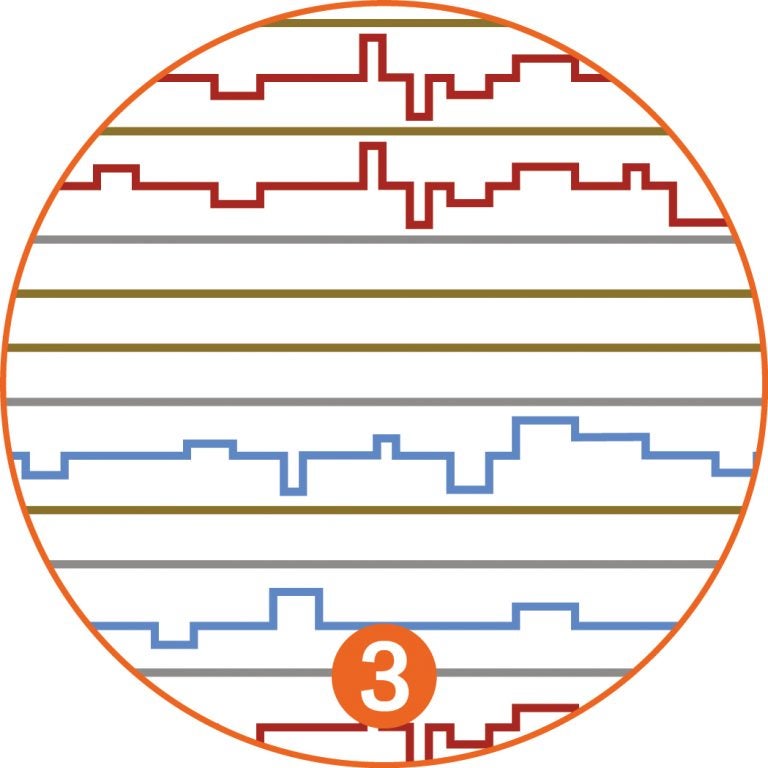
The biggest challenge, Krasnitz says, is correct analysis of cells that superficially look like tumor cells but are not cancerous. The look-alikes, dubbed UTLs (unrelated tumor-like cells—colored blue), have irregular CNV profiles that indicate chromosome irregularities not caused by cancer [3].
Krasnitz and colleagues ran their test making a very conservative assumption: that large numbers of UTL cells would survive the screen. The math is complex, but the team is satisfied that UTL cells ultimately will fail the test of clonality. They have irregular CNV profiles, but they don’t resemble one another (blue lines in [3]). Cancerous clonal cells have identical CNV profiles (red lines in [3]).
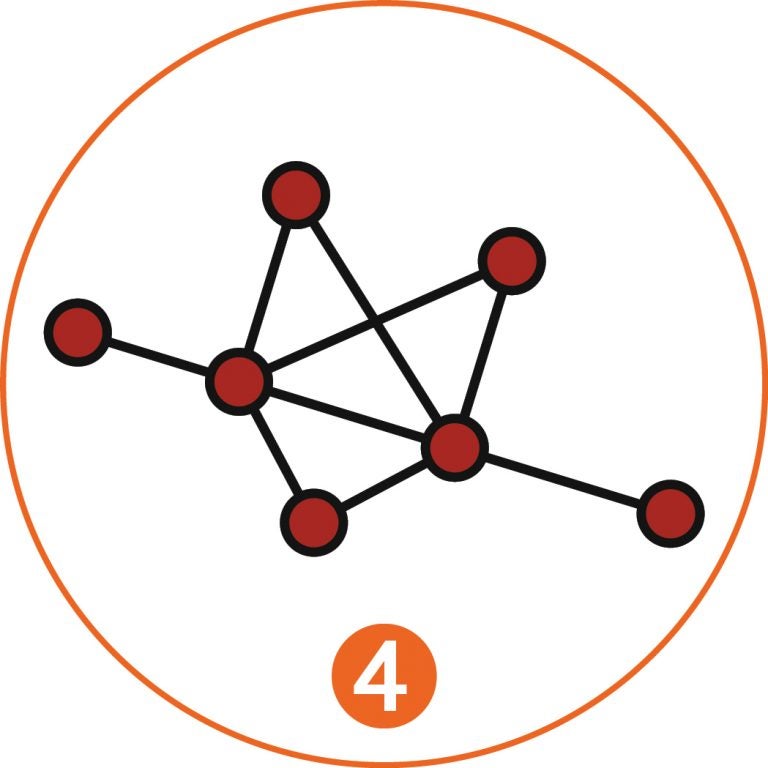
A mathematical procedure, or algorithm, is at the heart of making the crucial leap: finding similarities of pattern in the CNV profiles of suspicious cells in the filtered sample—what the team calls “pairwise correlations.” By following such relationships, constellations of related cells appear, and are mapped. More than six “connected components” [4] are what the team regards as a positive signal, indicating clonality and therefore a very strong possibility of cancer’s presence somewhere in the blood donor’s body. Follow-up analysis would identify the tissue of origin of the clonal cells, which would then be scanned at high resolution to pinpoint the incipient tumor.
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455