Study of two breast cancer samples suggests tumors grow by ‘punctuated, clonal expansions’
Cold Spring Harbor, NY — A new method of analyzing cancerous tumors developed by scientists at Cold Spring Harbor Laboratory (CSHL) suggests that tumors may not evolve gradually, but rather in punctuated or staccato-like bursts. It is a finding that has already shed new light on the process of tumor growth and metastasis, and may help in the development of new methods to clinically evaluate tumors.
The new analytic method, devised by CSHL Professor Michael Wigler and colleagues, features a process called single cell sequencing (SNS), which enables accurate quantification of genomic copy number within a single cell nucleus. Genomic copy number refers to the amount of DNA in the nucleus. In cancer, portions of the genome are amplified or deleted, giving rise to extra or missing copies of key genes and interfering with mechanisms that normally control cell growth.
In a study published today in the journal Nature, “we demonstrated that we can obtain accurate and high-resolution copy number profiles by sequencing a single cell from a cancerous tumor,” says Wigler, “and that by examining multiple cells from the same cancer, we can make inferences about how the cancer evolved and spread.” The CSHL team also includes Professor W. Richard McCombie, Assistant Professor Alex Krasnitz and Research Professor James Hicks. Nicholas Navin, the paper’s first author, was a graduate student while pursuing the research at CSHL and is now Assistant Professor at MD Anderson Cancer Center in Texas.
The importance of cancer’s heterogeneity
It has been very difficult for scientists to translate their growing ability to classify tumors at the molecular level into methods and tests that can be used in the clinic to analyze tumors in actual patients. But basic research has firmly established that cancer is highly heterogeneous, meaning that cancers of a certain type, for instance, cancers of the breast or lung or colon, can be subdivided into many distinct subtypes. Unique markers have been found in a number of cancer subtypes, such as abnormally high levels of the receptor protein encoded by the HER2neu gene, which in a subset of breast cancer cases is a causal factor in oncogenesis. The HER2neu receptor is the target for the pathbreaking drug Herceptin™ (Genentech Roche).
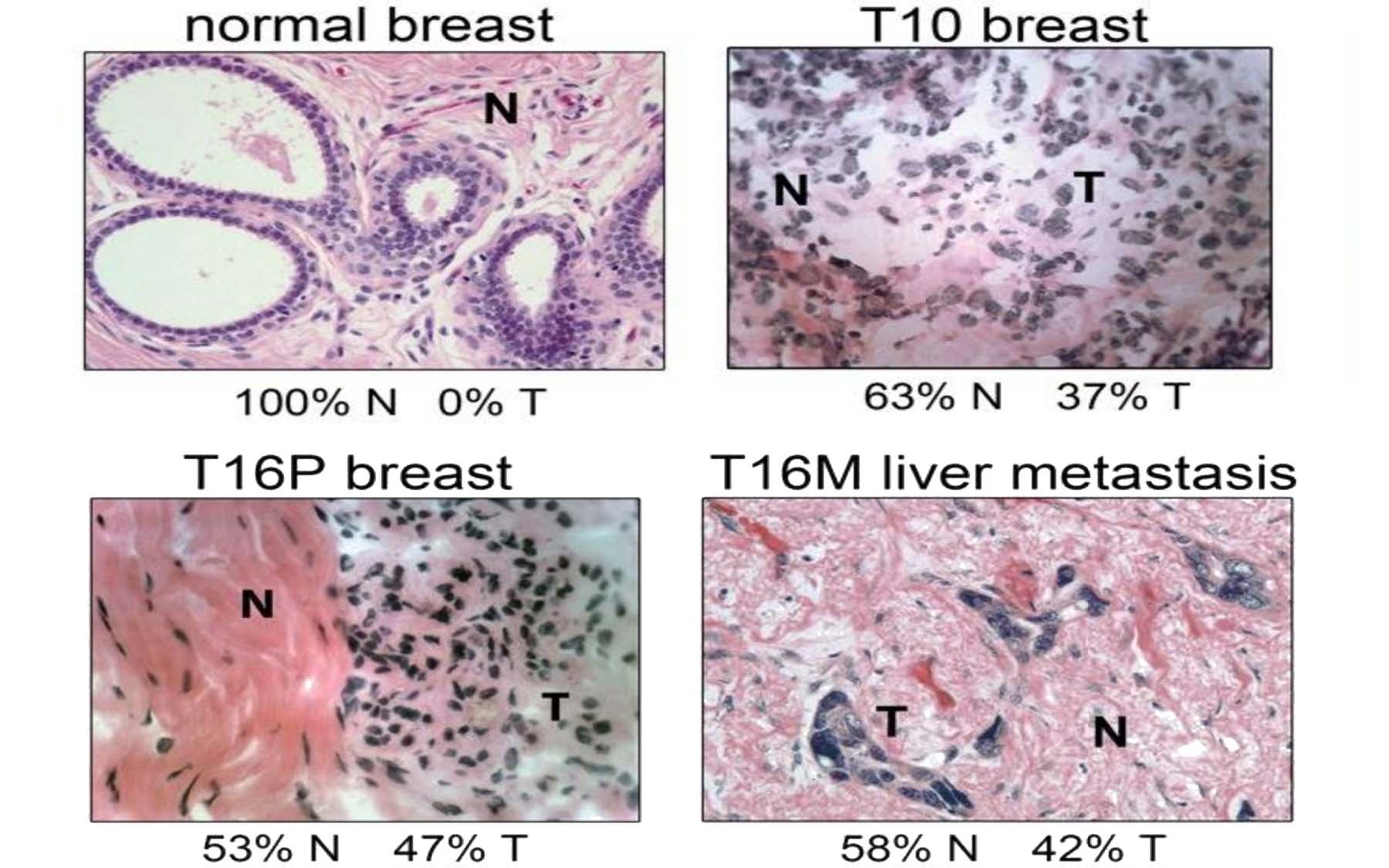
In the CSHL team’s just-published study, the two sampled tumors were known in advance—because of prior study—to represent distinct types. Both were primary invasive breast cancer tumors of the so-called “triple-negative” type, generally regarded as the most aggressive form of breast cancer. One tumor sample was known from prior testing to be polygenomic: composed of distinct populations of tumor cells, whose number, genomic type and evolutionary history were not readily measurable using conventional techniques. The other sampled tumor was monogenomic: composed of cells of a single genetic type. Unlike the first sample, this one had metastasized, to the liver, and samples of that tumor were also subjected to analysis.
Using SNS, in concert with whole-genome amplification and next-generation sequencing, the CSHL team was able to show that in fact three distinct subpopulations of tumor cells were present. “By SNS we could now see that each of these subpopulations was composed of cells that shared highly similar copy number profiles,” says Navin. “We could infer that these most probably represent three distinct clonal expansions of the tumor.”
Distinct clonal populations as evolutionary ‘winners’
Cancer has long been understood to be a clonal disease at the cellular level, with what in evolutionary terms might be called “successful” tumor cells providing the genetic template for the next generation of tumor-cell offspring. The cancer cells comprising the rapidly expanding malignant clone, it is thought, are those best able to survive in the hostile environment of the growing tumor, into which the body’s own immune cells have migrated and perhaps also toxic chemotherapy drugs have been introduced. This is one way of understanding why cancers are so tough to combat, and why they are able to survive even the application of cytotoxic therapies. In short, cancer cells are often able to evolve around these.
It has been theorized that genetically heterogeneous tumor cell populations become mixed via gradual evolution. The CSHL team, in contrast, suggests the evolution is likely punctuated. “It’s a very complicated matter to try to determine a given tumor’s genetic evolution,” says Hicks of the CSHL team. “The different groups of cells are geographically and anatomically intertwined. In evolutionary terms, you might think of them as distinct but intermixed ‘tribes,’ or subspecies. It’s important to keep in mind that tumors are chaotic places. There’s always a certain amount of chaotic evolution taking place, some of it reflected in rearrangement of chromosomes and some in single base-pair changes in the DNA of individual tumor cells. But our results, although based so far on a limited sample size, suggest that major clonal expansion events are relatively infrequent and may provide accessible targets for targeted treatments.”
Implications for understanding metastasis
The second breast cancer sample analyzed by the team was of a single genomic type, but the team was able to determine that the liver metastasis to which it had given rise was very closely related, in genomic terms. “The data suggest the primary tumor mass formed by a single clonal expansion and that one of the cells from this event subsequently seeded the metastatic tumor, with little further evolution,” according to Navin. “Although closely related, primary and metastatic tumor cells were cleanly separated, indicating to us that the two populations had not mixed” since the origin of the metastasis. Further, he noted, “differences in the profiles that distinguish primary and metastatic tumor populations were in the degree of copy number changes.”
A method called chromosomal breakpoint analysis was the means by which the team drew inferences about the relationships between various tumor cell subpopulations. “The history of the cell is written in these breakpoints,” explains Hicks. “They are the product of mutational events, the rearrangements of DNA that have occurred, and they don’t go away.” Generally speaking, these events are additive, a key concept that enabled the CSHL team to say that the genetic provenance of a cell is written in its DNA. The evolutionary history of the polygenomic tumor sample studied was traced by tracking 657 distinct chromosomal breakpoints. Tumor cells of all three clonal “subspecies” shared about half the mutational events; only a fraction shared another significant number of mutations; and a still smaller fraction shared yet another wave of mutations.
A significant fraction of tumor cells—up to 30%—contained some combination of DNA deletions and additions that made them appear normal, in terms of their total DNA content. These cells, called “pseudodiploid” by the researchers, appear to be cancerous but are not part of the major clonal expansions. The team’s method provides the first means of identifying such cells, which are not distinguishable using conventional methods of tumor characterization. One hypothesis holds that such cells are potential sources of future clonal events; another, that they may be important in the process of metastasis.
“We want to learn how a metastasis grows,” said Hicks, “and we now have the ability to do that at a very detailed level. In a typical blood draw taken from a cancer patient, you will typically see between five and 20 circulating tumor cells, a mark of metastasis. We’ll be able to look at every one of them and see if, for example, there’s a new clone that is different from the cell populations in the primary tumor. By working at a granular level, cell by cell, we can look deeply into a metastasis and see how it got built.”
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Citation
“Tumor Evolution Inferred by Single Cell Sequencing” appears online ahead of print in Nature on March 13, 2011. The authors are: Nicholas Navin, Jude Kendall, Jennifer Troge, Peter Andrews, Linda Rodgers, Jeanne McIndoo, Kerry Cook, Asya Stepansky, Dan Levy, Diane Esposito, Lakshmi Muthuswamy, Alex Krasnitz, Richard McCombie, James Hicks and Michael Wigler. The paper can be viewed online at: www.nature.com; the DOI is: 10.1038/nature09807