A theoretical physicist by training, my research is centered around intelligent machines. I do both theoretical and experimental work. The theoretical work is focussed on analyzing distributed/networked algorithms in the context of control theory and machine learning, using tools from statistical physics. My lab is involved in brain-wide mesoscale circuit mapping in the Mouse as well as in the Marmoset. An organizing idea behind my research is that there may be common underlying mathematical principles that constrain evolved biological systems and human-engineered systems.
Partha Mitra is interested in understanding intelligent machines that are products of biological evolution (particularly animal brains), with the basic hypothesis that common underlying principles may govern these “wet” intelligent machines and the “dry” intelligent machines that are transforming the present economy. Dr Mitra initiated the idea of brain-wide mesoscale circuit mapping, and his laboratory is involved in carrying out such mapping in the Mouse (http://mouse.brainarchitecture.org) and the Marmoset (in collaboration with Japanese and Australian scientists at the RIKEN Brain Science Institute and Monash University).
Dr Mitra spent ten years as a member of the theory department at Bell Laboratories and holds a visiting professorship at IIT Madras where he is helping establish the Center for Computational Brain Research. He has an active theoretical research program in machine learning and control theory, wheren he is using tools from statistical physics to analyze the performance of distributed/networked algorithms in the “thermodynamic” limit of many variables.
- Fellow, American Physical Society
- Senior Member, IEEE
- H N Mahabala Chair Professor (visiting), IIT Madras
- Partha Mitra awarded NSF ‘Early Concept’ grant for neuroscience
- CSHL’s Partha Mitra receives two awards for theoretical work with implications for brain circuitry
- Senior Visiting Scientist, RIKEN Brain Science Institute
- Mitra awarded “Transformative” NIH Grant
- George S. Axelby Outstanding Paper Award
NeuroAI with an eye on equity
September 30, 2024
Working at the intersection of neuroscience and computer science, CSHL researchers aim to build AI that will benefit everyone, not just the lucky few.
Mitra among first awarded NIH BRAIN CONNECTS grant
September 26, 2023
New National Institutes of Health initiative aims to generate an atlas of brain connections, offering new insights into neurological disorders.
Laying the groundwork for drug discoveries
August 8, 2023
A new partnership between CSHL and one of the world’s leading biotech investors could streamline this process and help change society for the better.
Building on 150 years of neuroanatomy
October 7, 2021
Learn more about how researchers reached a milestone in a years-long effort to catalog the cells of the human, mouse, and monkey brains.
Think a census of humans is hard? Try counting their brain cells!
October 6, 2021
CSHL researchers and other collaborators reached a milestone in a years-long effort to catalog the cells of the human, mouse, and monkey brains.
Research matters
June 8, 2021
Innovative research and educational activities never stopped during the COVID-19 pandemic.
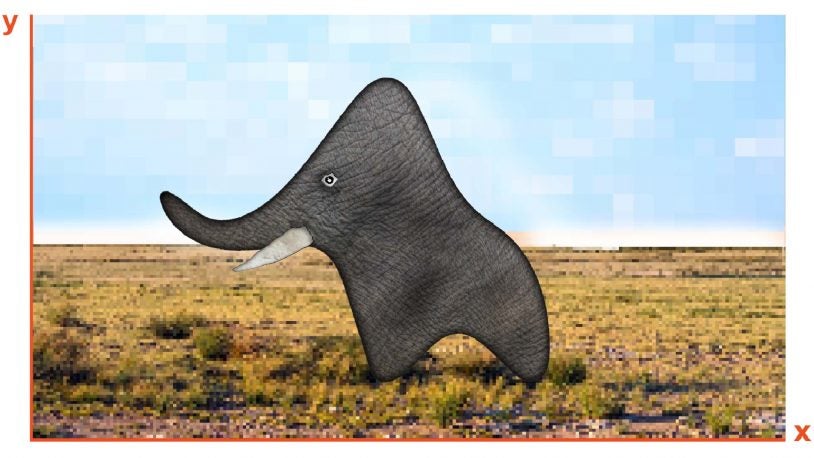
Let’s talk about the elephant in the data
June 3, 2021
How much prior knowledge does a machine learning computer need to find the truth? CSHL Professor Partha Mitra looks to human brains for an answer.
NIH BRAIN Initiative invests $9.7 million in CSHL scientists
December 29, 2020
CSHL scientists received grants to broaden our knowledge of the human brain and how to treat neurological disorders.
Partha Mitra: AI and brain circuits
November 30, 2020
Mitra answers audience questions about how understanding brain connections can bring medical and technological breakthroughs.
LIVE at the Lab with Partha Mitra: AI and brain circuits
November 27, 2020
CSHL Professor Partha Mitra presents some challenges for neuroscientists and how artificial intelligence (AI) helps overcome them.
Selected Publications
Fitting elephants in modern machine learning by statistically consistent interpolation
1 May 2021 | Nature Machine Intelligence | 3(5):378-386
Mitra, P;
A multimodal cell census and atlas of the mammalian primary motor cortex
21 Oct 2020 | bioRxiv
BRAIN Initiative Cell Census Network (BICCN); Adkins, Ricky; Aldridge, Andrew; Allen, Shona; Ament, Seth; An, Xu; Armand, Ethan; Ascoli, Giorgio; Bakken, Trygve; Bandrowski, Anita; Banerjee, Samik; Barkas, Nikolaos; Bartlett, Anna; Bateup, Helen; Behrens, Margarita; Berens, Philipp; Berg, Jim; Bernabucci, Matteo; Bernaerts, Yves; Bertagnolli, Darren; Biancalani, Tommaso; Boggeman, Lara; Booeshaghi, Sina; Bowman, Ian; Bravo, Héctor; Cadwell, Cathryn; Callaway, Edward; Carlin, Benjamin; O'Connor, Carolyn; Carter, Robert; Casper, Tamara; Castanon, Rosa; Castro, Jesus; Chance, Rebecca; Chatterjee, Apaala; Chen, Huaming; Chun, Jerold; Colantuoni, Carlo; Crabtree, Jonathan; Creasy, Heather; Crichton, Kirsten; Crow, Megan; D'Orazi, Florence; Daigle, Tanya; Dalley, Rachel; Dee, Nick; Degatano, Kylee; Dichter, Benjamin; Diep, Dinh; Ding, Liya; Ding, Song-Lin; Dominguez, Bertha; Dong, Hong-Wei; Dong, Weixiu; Dougherty, Elizabeth; Dudoit, Sandrine; Ecker, Joseph; Eichhorn, Stephen; Fang, Rongxin; Felix, Victor; Feng, Guoping; Feng, Zhao; Fischer, Stephan; Fitzpatrick, Conor; Fong, Olivia; Foster, Nicholas; Galbavy, William; Gee, James; Ghosh, Satrajit; Giglio, Michelle; Gillespie, Thomas; Gillis, Jesse; Goldman, Melissa; Goldy, Jeff; Gong, Hui; Gou, Lin; Grauer, Michael; Halchenko, Yaroslav; Harris, Julie; Hartmanis, Leonard; Hatfield, Joshua; Hawrylycz, Mike; Helba, Brian; Herb, Brian; Hertzano, Ronna; Hintiryan, Houri; Hirokawa, Karla; Hockemeyer, Dirk; Hodge, Rebecca; Hood, Greg; Horwitz, Gregory; Hou, Xiaomeng; Hu, Lijuan; Hu, Qiwen; Huang, Josh; Huo, Bingxing; Ito-Cole, Tony; Jacobs, Matthew; Jia, Xueyan; Jiang, Shengdian; Jiang, Tao; Jiang, Xiaolong; Jin, Xin; Jorstad, Nikolas; Kalmbach, Brian; Kancherla, Jayaram; Keene, Dirk; Kelly, Kathleen; Khajouei, Farzaneh; Kharchenko, Peter; Kim, Gukhan; Ko, Andrew; Kobak, Dmitry; Konwar, Kishori; Kramer, Daniel; Krienen, Fenna; Kroll, Matthew; Kuang, Xiuli; Kuo, Hsien-Chi; Lake, Blue; Larsen, Rachael; Lathia, Kanan; Laturnus, Sophie; Lee, Angus; Lee, Cheng-Ta; Lee, Kuo-Fen; Lein, Ed; Lesnar, Phil; Li, Anan; Li, Xiangning; Li, Xu; Li, Yang; Li, Yaoyao; Li, Yuanyuan; Lim, Byungkook; Linnarsson, Sten; Liu, Christine; Liu, Hanqing; Liu, Lijuan; Lucero, Jacinta; Luo, Chongyuan; Luo, Qingming; Macosko, Evan; Mahurkar, Anup; Martone, Maryann; Matho, Katherine; McCarroll, Steven; McCracken, Carrie; McMillen, Delissa; Miranda, Elanine; Mitra, Partha; Miyazaki, Paula; Mizrachi, Judith; Mok, Stephanie; Mukamel, Eran; Mulherkar, Shalaka; Nadaf, Naeem; Naeemi, Maitham; Narasimhan, Arun; Nery, Joseph; Ng, Lydia; Ngai, John; Nguyen, Thuc; Nickel, Lance; Nicovich, Philip; Niu, Sheng-Yong; Ntranos, Vasilis; Nunn, Michael; Olley, Dustin; Orvis, Joshua; Osteen, Julia; Osten, Pavel; Owen, Scott; Pachter, Lior; Palaniswamy, Ramesh; Palmer, Carter; Pang, Yan; Peng, Hanchuan; Pham, Thanh; Pinto-Duarte, Antonio; Plongthongkum, Nongluk; Poirion, Olivier; Preissl, Sebastian; Purdom, Elizabeth; Qu, Lei; Rashid, Mohammad; Reed, Nora; Regev, Aviv; Ren, Bing; Ren, Miao; Rimorin, Christine; Risso, Davide; Rivkin, Angeline; Muñoz-Castañeda, Rodrigo; Romanow, William; Ropelewski, Alexander; Roux de Bézieux, Hector; Ruan, Zongcai; Sandberg, Rickard; Savoia, Steven; Scala, Federico; Schor, Michael; Shen, Elise; Siletti, Kimberly; Smith, Jared; Smith, Kimberly; Somasundaram, Saroja; Song, Yuanyuan; Sorensen, Staci; Stafford, David; Street, Kelly; Sulc, Josef; Sunkin, Susan; Svensson, Valentine; Tan, Pengcheng; Tan, Zheng; Tasic, Bosiljka; Thompson, Carol; Tian, Wei; Tickle, Timothy; Tieu, Michael; Ting, Jonathan; Tolias, Andreas; Torkelson, Amy; Tung, Herman; Vaishnav, Eeshit; Van den Berge, Koen; van Velthoven, Cindy; Vanderburg, Charles; Veldman, Matthew; Vu, Minh; Wakeman, Wayne; Wang, Peng; Wang, Quanxin; Wang, Xinxin; Wang, Yimin; Wang, Yun; Welch, Joshua; White, Owen; Williams, Elora; Xie, Fangming; Xie, Peng; Xiong, Feng; Yang, William; Yanny, Anna; Yao, Zizhen; Yin, Lulu; Yu, Yang; Yuan, Jing; Z, Hongkui;
Cellular Anatomy of the Mouse Primary Motor Cortex
2 Oct 2020 | bioRxiv
Muñoz-Castañeda, Rodrigo; Zingg, Brian; Matho, Katherine; Wang, Quanxin; Chen, Xiaoyin; Foster, Nicholas; Narasimhan, Arun; Li, Anan; Hirokawa, Karla; Huo, Bingxing; Bannerjee, Samik; Korobkova, Laura; Park, Chris; Park, Young-Gyun; Bienkowski, Michael; Chon, Uree; Wheeler, Diek; Li, Xiangning; Wang, Yun; Kelly, Kathleen; An, Xu; Attili, Sarojini; Bowman, Ian; Bludova, Anastasiia; Cetin, Ali; Ding, Liya; Drewes, Rhonda; D’Orazi, Florence; Elowsky, Corey; Fischer, Stephan; Galbavy, William; Gao, Lei; Gillis, Jesse; Groblewski, Peter; Gou, Lin; Hahn, Joel; Hatfield, Joshua; Hintiryan, Houri; Huang, Jason; Kondo, Hideki; Kuang, Xiuli; Lesnar, Philip; Li, Xu; Li, Yaoyao; Lin, Mengkuan; Liu, Lijuan; Lo, Darrick; Mizrachi, Judith; Mok, Stephanie; Naeemi, Maitham; Nicovich, Philip; Palaniswamy, Ramesh; Palmer, Jason; Qi, Xiaoli; Shen, Elise; Sun, Yu-Chi; Tao, Huizhong; Wakemen, Wayne; Wang, Yimin; Xie, Peng; Yao, Shenqin; Yuan, Jin; Zhu, Muye; Ng, Lydia; Zhang, Li; Lim, Byung; Hawrylycz, Michael; Gong, Hui; Gee, James; Kim, Yongsoo; Peng, Hanchuan; Chuang, Kwanghun; Yang, William; Luo, Qingming; Mitra, Partha; Zador, Anthony; Zeng, Hongkui; Ascoli, Giorgio; Huang, Josh; Osten, Pavel; Harris, Julie; Dong, Hong-Wei;
Semantic segmentation of microscopic neuroanatomical data by combining topological priors with encoder-decoder deep networks
Oct 2020 | Nature Machine Intelligence | 2(10):585-+
Banerjee, Samik; Magee, Lucas; Wang, Dingkang; Li, Xu; Huo, Bing-Xing; Jayakumar, Jaikishan; Matho, Katherine; Lin, Meng-Kuan; Ram, Keerthi; Sivaprakasam, Mohanasankar; Huang, Josh; Wang, Yusu; Mitra, Partha;
ZEBrA: Zebra finch Expression Brain Atlas-A resource for comparative molecular neuroanatomy and brain evolution studies
Aug 2020 | Journal of Comparative Neurology | 528(12):2099-2131
Lovell, P; Wirthlin, M; Kaser, T; Buckner, A; Carleton, J; Snider, B; McHugh, A; Tolpygo, A; Mitra, P; Mello, C;
All Publications
Skeletonization of neuronal processes using Discrete Morse techniques from computational topology
15 May 2025 | bioRxiv
Banerjee, Samik; Stam, Caleb; Tward, Daniel; Savoia, Steven; Wang, Yusu; Mitra, Partha;
3D multimodal histological atlas and coordinate framework for the mouse brain and head
14 May 2025
Mitra, Partha; Flannery, Patrick; Savoia, Stephen; Mezias, Christopher; Bannerjee, Samik; Richman, Max; Lodato, Brianna; O'Rourke, Joseph; Balani, Somesh; Arima, Ken; Washington, Stuart; Coronado-Leija, Ricardo; Zhang, Jiangyang; Tward, Daniel;
What makes a theory of consciousness unscientific?
Apr 2025 | Nature Neuroscience | 28(4):689-693
IIT-Concerned; Klincewicz, Michał; Cheng, Tony; Schmitz, Michael; Sebastián, Miguel; Snyder, Joel;
DHARANI: A 3D Developing Human-Brain Atlas Resource to Advance Neuroscience Internationally Integrated Multimodal Imaging and High-Resolution Histology of the Second Trimester
Feb 2025 | Journal of Comparative Neurology | 533(2):e70006
Verma, Richa; Bota, Mihail; Ram, Keerthi; Jayakumar, Jaikishan; Folkerth, Rebecca; Pandurangan, Karthika; Ramesh, Jivitha; Majumder, Moitrayee; Raveendran, Rakshika; Nanda, Reetuparna; K, Sivamani; S, Amal; Karthik, Srinivasa; Kumarasami, Ramdayalan; S, Suresh; Lata, S; Kumar, E; Rangasami, Rajeswaran; Srinivasan, Chitra; Kumutha, Jayaraman; Vasudevan, Sudha; Bhat, Koushik; Sam C, Chrisline; Neelakantan, Sivathanu; Savoia, Stephen; Mitra, Partha; Joseph, Jayaraj; Manger, Paul; Sivaprakasam, Mohanasankar;
A three-dimensional histological cell atlas of the developing human brain
10 Jan 2025
Mitra, Partha; Jayakumar, Jaikishan; Sivaprakasam, Mohanasankar; Verma, Richa; Bota, Mihail; Joseph, Jayaraj; Mulay, Supriti; Kumutha, Jayaraman; Sriniva, Chitra; S, Suresh; S, Latha; Kumar, Harish; Bhaduri, Aparna; Nowakowski, Tomasz; Roy, Prasun; Savoia, Stephen; Bannerjee, Samik; Tward, Daniel;