Read about CSHL’s contributions to neuroanatomy in recent years, and learn more about related milestones in our interactive timeline.
How does the human brain work? For the last 150 years, researchers have tried to assess its function from its structure, but the answer is still elusive. On October 7, 2021, scientists reached a major milestone, publishing a comprehensive catalog of cell types in the mouse, monkey, and human primary motor cortex in Nature.
The catalog culminates eight years of focused investment in neuroanatomy innovation. In 2013, the U.S. government launched a $100 million program aimed at understanding human brain anatomy. The large collaborative project was dubbed the Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative. After three years of developing computational and imaging tools, BRAIN Initiative neuroscientists and funders met at the CSHL Banbury Conference Center to decide on the best path forward. Geneticists had met two decades earlier in the same place to kick off one of the first large international biology collaborations: the Human Genome Project. The BRAIN Initiative stakeholders decided to take a census of every cell in the brain and figure out how cells connected to each other, an effort titled the BRAIN Initiative Cell Census Network (BICCN). Collaborators would examine small sections of brains in great detail, then scale up and study ever-larger portions.
A consortium of collaborators
One significant accomplishment for the BICCN was to gather a consortium of scientists from many fields, including neuroscientists, computational scientists, physicists, geneticists, and instrument makers. At the start of the initiative, CSHL Associate Professor Pavel Osten, former CSHL Professor Gregory Hannon, CSHL Professor Partha Mitra, and CSHL Associate Professor Dinu Florin Albeanu (funded by the National Institutes of Health and the National Science Foundation) developed computational and imaging tools.

Z. Josh Huang, then a professor at CSHL, recalls that a hundred years earlier, researchers studied the shapes and organization of cells in the brain. Later, others used human functional MRI to measure brain activity during certain behavioral tasks. Neither method had the resolution to understand how neural circuits truly work at the speed of thought. According to Huang:
“Ultimately, the brain computes through individual cells, communicating to each other at the level of synapses. The brain, of course, is a sophisticated neural network or computational machine. So, to understand any kind of network, you need to first understand the basic components and how they are connected.”
Developments across many fields allowed scientists to discover and characterize cell types in different ways. Mitra says:
“Neuroscience has not been big science. It has been little science, individual investigator-driven research. So I would say the BICCN is one of the most coherent, and should I say, successful consortia, that exists within the BRAIN Initiative. Creating these resources, it’s challenging, unglamorous work. What is interesting is that scientists are willing to come together to do this kind of unglamorous work. And the sum is greater than the parts as a result.”
How to build a neuroanatomy revolution
It takes more than money and people to make a revolution; it takes a clear set of goals. A generation ago, the Human Genome Project defined toolkits and standards to spark a genomics revolution, eventually allowing anyone to sequence key parts of their genome and compare it to 30 million other humans for less than $100. Similarly, the BICCN created the tools and standards upon which all neuroanatomists will build going forward. The technology developed to understand a section of the brain will now be applied to counting and mapping every cell in the human brain, a structure far larger than anything examined before.
The goals of the BICCN are to:
- generate a census of each cell type and its locations, then develop tools to target these cell types.
- create a high-resolution atlas of cell types in the mouse brain and connectivity maps of other brain areas.
- create an atlas of human brain cell types from development to adult to study diseases.
- profile cell types in non-human primates, spanning the evolutionary gap between the mouse and human brain.
The atlases and catalogs published by the BICCN so far are meant to be frameworks. In July 2022, the BRAIN Initiative will start the next phase called the BRAIN Initiative Cell Atlas Network (BICAN), funded with a grant from the National Institutes of Health. Researchers will build cell-by-cell atlases of human and non-human primates. Neuroanatomists will be able to compare the human brain to the brains of other species important for research. Researchers hope that within the next ten years, thousands of human brains will be mapped and incorporated into reference atlases. This knowledge could be harnessed to study and treat schizophrenia, depression, Alzheimer’s, and traumatic brain injuries, and will revolutionize neuroscience as a whole.
CSHL contributions
Five principal investigators at CSHL conducted key BICCN and BRAIN Initiative projects. Follow the links to read more about each of their contributions.
Jesse Gillis (CSHL Associate Professor): Guilt by association
CSHL Associate Professor Jesse Gillis and his lab developed a computer tool to categorize cells based on similarities in their component parts. This statistics-based program, called MetaNeighbor, uses RNA transcripts (the instructions for the components) and uncovers which transcripts work together. It finds these relationships by assuming “guilt by association:” if two transcripts act similarly from cell to cell, then they likely relate to similar functions in both. Using a panel of transcripts covering many different functions, MetaNeighbor compared and categorized mammalian brain cells for the BICCN. It combined data from different labs that are using different methods into one comprehensive data set.
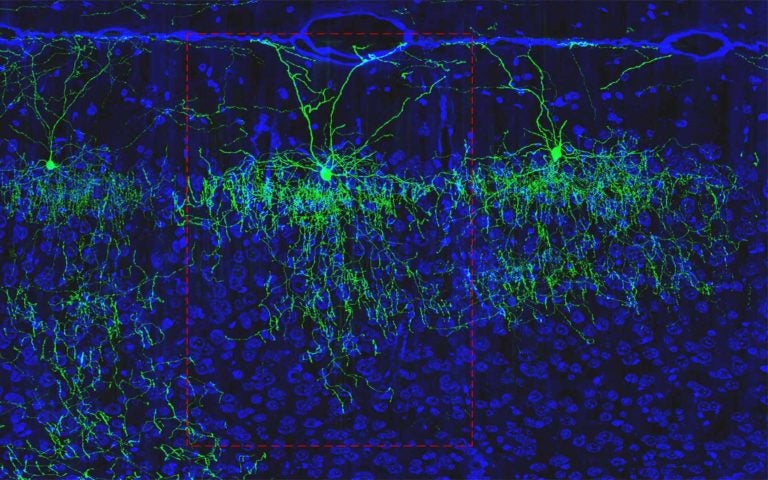
Z. Josh Huang (CSHL Adjunct Professor): Classifying brain cells
Within the BICCN, Huang leads the Comprehensive Center for Mouse Brain Atlas, which includes five principal investigators from CSHL and researchers from several other institutions. He has outlined ways to classify new cell subtypes based on their shapes, connections with other neurons, and the genes they use. The team is collecting anatomic and molecular information within the mouse forebrain. Huang was just awarded a prestigious NIH Director’s Pioneer Award to develop a new generation of precise and customizable cell engineering technologies. Researchers will be able to monitor and edit the function of diverse cell types across animal organs and species, with potentially broad applications in biomedical research, biotechnology, and therapeutics.
Partha Mitra (CSHL Professor): Building a computer that could map the brain
Mitra and other CSHL collaborators dramatically improved the efficiency of automated methods for tracing neuronal connections. They taught a computer to recognize different parts of neurons, then mapped the cells onto a topological world to see how those neurons are likely to connect. Such connection maps are critical for learning how the brain processes information to generate thoughts and behavior.
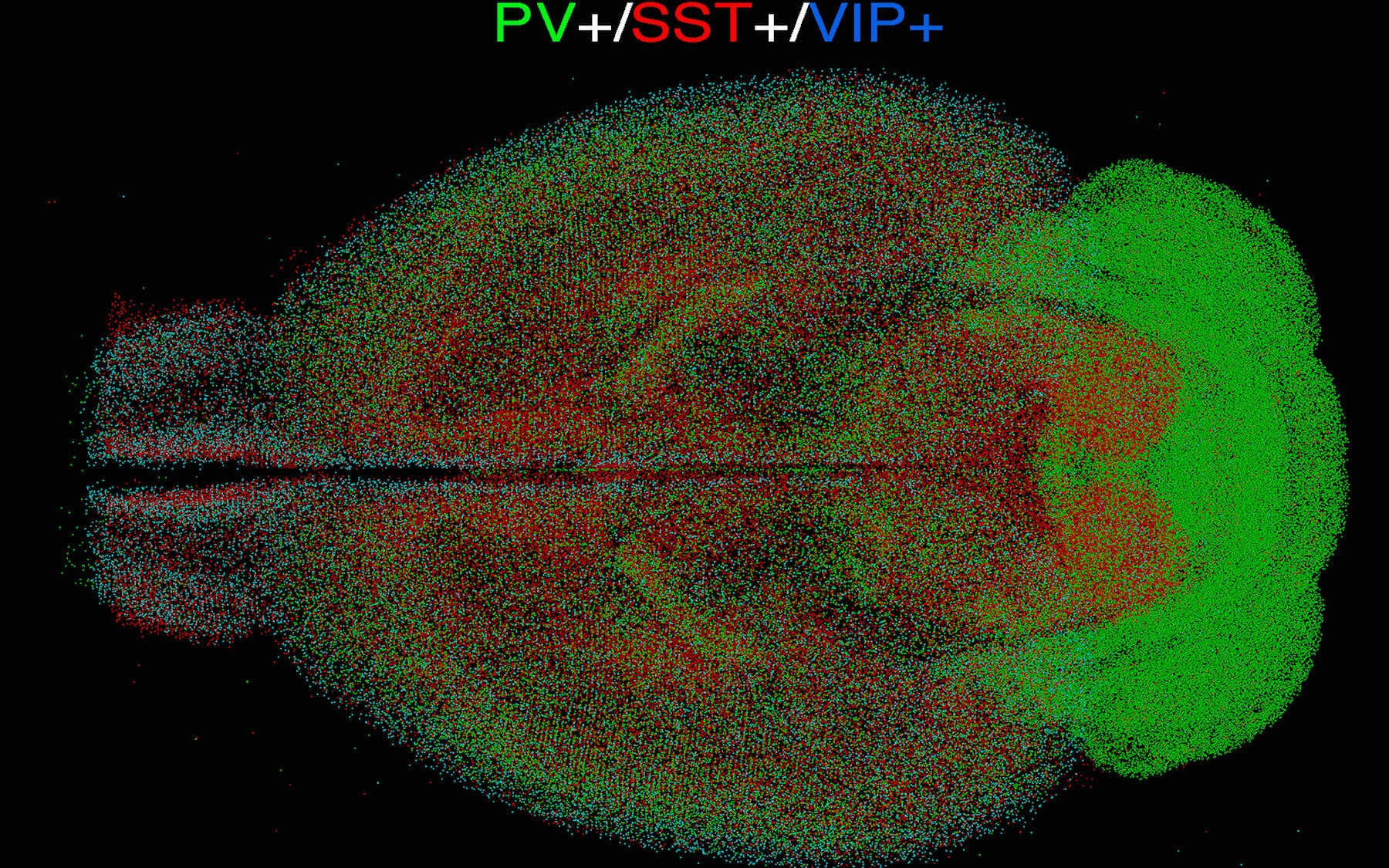
Pavel Osten (CSHL Associate Professor): Standardizing brains
Osten leads the Collaboratory for Atlasing Cell Type Anatomy in the Female and Male Mouse Brain, another branch of the BICCN dedicated to establishing cell-type distribution in the female and male mouse brains. He and his lab developed qBrain, a method that combines various brain imaging techniques, to map cells and connections of the mouse primary motor cortex in three dimensions. However, making observations on a single brain, a so-called “n=1”, is not sufficient to understand what is typical in a population of brains. Osten says, “100 brains would be a start, though 1,000 would be better,” noting:
“If you’re making an observation on ‘n=1’, you don’t know what’s important and what’s not because you just have an n=1 of everything.”
With many observations, Osten says, “you can start learning about the variability in the human brain, human gene expression. What is stereotypical, what is noise? And that’s sort of the critical part.” With a large amount of data, researchers will be able to tease out what differs between males and females, young and old, diseased and non-diseased states.
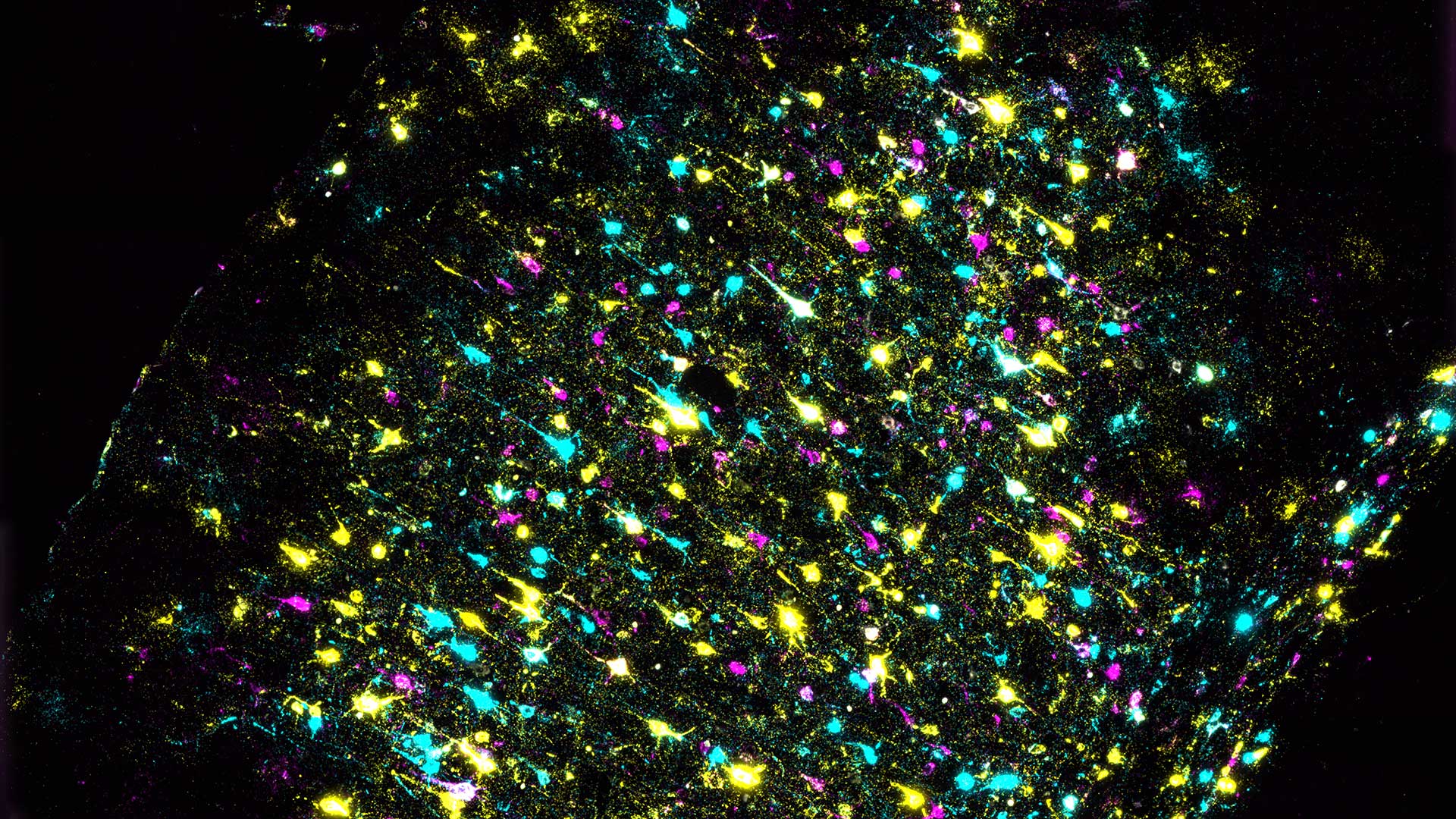
Anthony Zador (CSHL Professor): Barcoding is not just for retail stores
CSHL Professor Anthony Zador and his lab developed MAPseq, used to map the connectivity of different brain cells and to gain a better understanding of how they interact with one another. The technique labels brain cells with short RNA sequences, allowing researchers to trace individual neurons. Several years later, Zador and his team developed the next generation, BARseq, which can map thousands of neurons in a single mouse at single-neuron resolution. The latest update, BARseq2, adds the ability to determine connectivity and gene use in the same neurons.
Written by: Jasmine Lee, Content Developer/Communicator | publicaffairs@cshl.edu | 516-367-8845