Lucas Martin Cheadle
Associate Professor
Cancer Center Member
Ph.D., Neuroscience, Yale University, 2014
cheadle@cshl.edu | 516-367-5920
The trillions of connections between brain cells enable complex thought and behavior. These connections are wired with great precision through both genetics and in response to an organism’s experiences. Our lab seeks to understand how experiences engage specialized immune cells called microglia to shape the connectivity and function of the brain. We are further interested in how impairments in these processes can contribute to neurodevelopmental disorders such as autism.
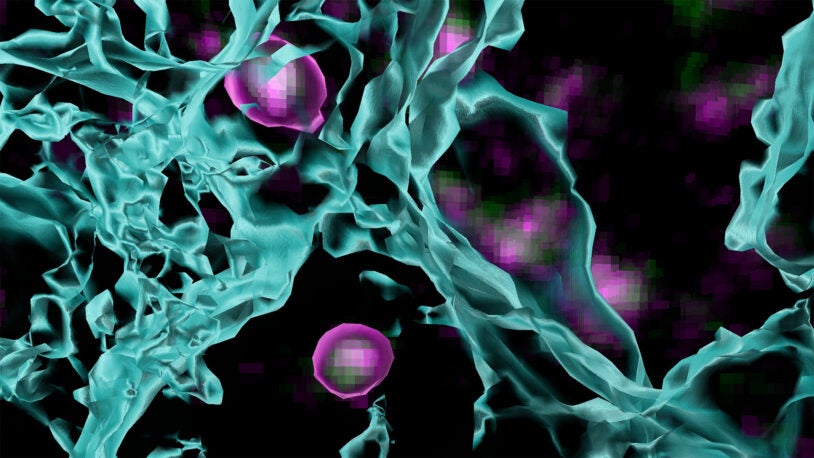
The powerful influence of sensory experience on brain development has been appreciated since the 1960s. Yet, even today, the fundamental cellular and molecular mechanisms through which sensory input shapes developing neural circuits remain largely mysterious. The Cheadle lab recently discovered that sensory experience alters gene and protein expression in microglia, the resident immune cells of the brain. These sensory-induced changes allow microglia to interact with neighboring neurons to strengthen and maintain a subset of synaptic connections and to eliminate others. These findings raise the exciting possibility that microglia, which are predominantly associated with immune responses to injury and disease, also decode salient features of the physical world and contribute to neural responses to the environment.
The Cheadle lab applies a multidisciplinary approach to the visual system of the mouse to investigate the contributions of microglia to sensory experience-dependent synapse development and plasticity. They further seek to identify the molecular mechanisms through which microglia effect changes at synapses and thereby exert control over brain function. To accomplish this, the Cheadle lab images microglial interactions with synapses in the brains of living mice, which allows the researchers to characterize the specific features of the environment to which microglia respond. In parallel, the research team uses cutting-edge single-cell transcriptomic and genomic strategies, such as single-cell RNA-sequencing, to profile the molecular changes in microglia that are elicited by distinct sensory stimuli. With these combined approaches, the Cheadle lab is interrogating the ways in which environmental stimuli converge upon the microglial genome to shape neural circuit development and function.
The CSHL School of Biological Sciences’ class of 2025
May 5, 2025
The School of Biological Sciences awarded Ph.D. degrees to nine students this year. Read some of their stories and reflections on their time at CSHL.
At the Lab: Products of an unseen environment
April 22, 2025
You may have read about their fascinating research in
Lucas Cheadle awarded $750K MIND Prize
April 1, 2025
Support from the Pershing Square Foundation empowers the CSHL neuroscientist to take a bold, new view of Alzheimer’s disease.
One Experiment: The brain’s landscapers
October 10, 2024
The brain relies on cells called OPCs to refine neural connections. CSHL’s Lucas Cheadle can now look at these synapse pruners in a whole new light.
CSHL postdoc presents breakthrough autism findings
October 7, 2024
Irene Sanchez Martin’s research on prenatal inflammation’s role in autism is showcased at the Society for Neuroscience’s annual meeting.
Cocktails & Chromosomes: Immune cells’ brainy convos
May 20, 2024
CSHL Assistant Professor Lucas Cheadle invites you to listen in to the ongoing conversation between your immune system and your brain.
Hazen Tower
April 25, 2024
The Italian-style bell tower anchors CSHL’s Neuroscience Center. Its bell has rung out every hour on the hour, from 8 a.m. to 8 p.m., since 1991.
Lucas Cheadle named HHMI Freeman Hrabowski Scholar
May 9, 2023
Cheadle was selected for his leadership in neuroscience research and advancing diversity, equity, and inclusion in science.
Diversity drives Science Forward
April 20, 2023
CSHL hosts the first-ever Science Forward symposium, a two-day event for early-career researchers from historically marginalized groups.
Lucas Cheadle named a 2022 NIH Director’s New Innovator
October 4, 2022
Cheadle is CSHL’s first recipient of the NIH Director’s New Innovator Award. It supports his new work on inflammation and autism spectrum disorders.
All Publications
The Many Lives of an Oligodendrocyte Precursor Cell
15 Apr 2025 | Annual Review of Neuroscience
Buchanan, JoAnn; Cheadle, Lucas;
A single-cell transcriptomic atlas of sensory-dependent gene expression in developing mouse visual cortex
28 Feb 2025 | Development | :dev.204244
Xavier, Andre; Lin, Qianyu; Kang, Chris; Cheadle, Lucas;
High-confidence and high-throughput quantification of synapse engulfment by oligodendrocyte precursor cells
3 Oct 2024 | Nature Protocols
Kahng, Jessica; Xavier, Andre; Ferro, Austin; Tang, Samantha; Auguste, Yohan; Cheadle, Lucas;
A brain-body feedback loop driving HPA-axis dysfunction in breast cancer
14 Sep 2024 | bioRxiv
Gomez, Adrian; Wu, Yue; Zhang, Chao; Boyd, Leah; Wee, Tse-Luen; Gewolb, Joseph; Amor, Corina; Cheadle, Lucas; Borniger, Jeremy;
A single-cell transcriptomic atlas of sensory-dependent gene expression in developing mouse visual cortex
26 Jun 2024 | bioRxiv
Xavier, Andre; Lin, Qianyu; Kang, Chris; Cheadle, Lucas;
Disruption of Cholinergic Retinal Waves Alters Visual Cortex Development and Function
15 Apr 2024 | bioRxiv
Burbridge, Timothy; Ratliff, Jacob; Dwivedi, Deepanjali; Vrudhula, Uma; Alvarado-Huerta, Francisco; Sjulson, Lucas; Ibrahim, Leena; Cheadle, Lucas; Fishell, Gordon; Batista-Brito, Renata;
The cytokine receptor Fn14 is a molecular brake on neuronal activity that mediates circadian function in vivo
2 Apr 2024 | bioRxiv
Ferro, Austin; Arshad, Anosha; Boyd, Leah; Stanley, Tess; Berisha, Adrian; Vrudhula, Uma; Gomez, Adrian; Borniger, Jeremy; Cheadle, Lucas;
Integrated high-confidence and high-throughput approaches for quantifying synapse engulfment by oligodendrocyte precursor cells
25 Aug 2023 | bioRxiv
Kahng, Jessica; Xavier, Andre; Ferro, Austin; Auguste, Yohan; Cheadle, Lucas;
Emerging roles of oligodendrocyte precursor cells in neural circuit development and remodeling
Aug 2023 | Trends in Neurosciences | 46(8):628-639
Buchanan, JoAnn; da Costa, Nuno; Cheadle, Lucas;
Neuron-glia communication through bona fide synapses
5 May 2023 | Nature Reviews Neuroscience
Cheadle, Lucas;