Cold Spring Harbor Laboratory Professor Anthony Zador has taken the next step in his quest to solve exactly how the brain is wired.
Zador, a neuroscientist whose lab studies how the brain’s circuitry mediates and controls complex behaviors, set out about 10 years ago to map three pillars of brain function: connectivity, gene expression and physiological activity. Because the technology didn’t exist to effectively do that, his team developed MAPseq, a technique used to map the connections of different brain cells and gain a better understanding of how they interact with each other. Over the years, Zador and his lab continued to improve the technique.
In a study published in the latest issue of Cell, Zador’s lab, led by Xiaoyin Chen, a postdoctoral fellow and the first author on the paper, introduces BARseq, the next generation of MAPseq. The new technology can be used to expand the brain map by accurately pinpointing the location of a neuron. This allows BARseq to determine not only a neuron’s connections, but also its pattern of gene expression and its physiological activity, two pieces of the puzzle MAPseq couldn’t handle.
“The brain is basically a circuit. It’s a bunch of neurons connected to one another,” Zador said. “In order to make progress in neuroscience, we have to actually be able to understand how those neurons are connected to one another, and ideally to be able to relate them to other aspects of neural function, like gene expression and neuronal activity.”
BARseq is designed to help researchers move closer to that goal. It’s based on the same concept as MAPseq, with several important upgrades.
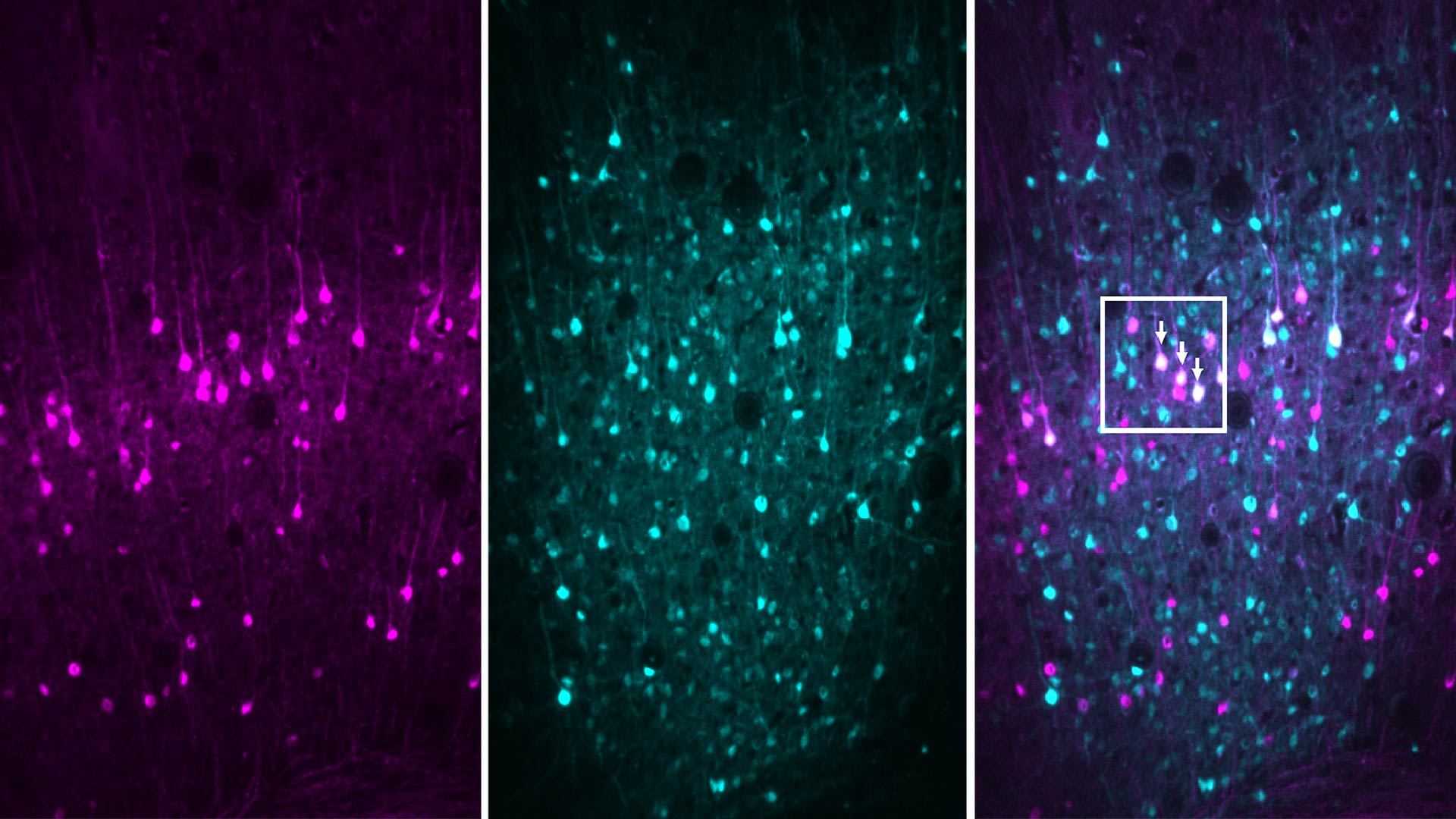
The researchers used MAPseq to tag each neuron with a unique barcode comprised of genetic sequences. By following those tags across the brain, they can see where the neurons send messages and then map the pathways those signals form between different areas of the brain. This is called brain connectivity.
The problem with MAPseq was that the process of tagging the neurons—where the brain cells had to be mixed together, separated and then tagged—resulted in poor spatial resolution. The process produced a blurry map that made it difficult for the researchers to see how a neuron was connected to a particular pillar, such as gene expression. The low resolution also prevented researchers from pinpointing exactly where in the brain the neuron was located.
BARseq solves that problem by allowing researchers to tag and sequence the neurons in situ, or in their original form and location on the brain. “So basically, you can see exactly where the neurons are when we sequence the barcodes,” said Chen.
The team used BARseq to map the connections of 3,579 neurons in the auditory cortex of the mouse brain. Matching connectivity patterns to gene expression allows scientists to characterize different cell types and define their specific functions in the brain. It could prove to be a valuable tool for studying how neural circuits are formed.
“If we could have a complete wiring diagram, a map, that would provide a foundation for understanding thought, consciousness, decision making and how those go awry in neuropsychiatric disorders like autism, schizophrenia, depression, OCD,” Zador said.
BARseq, like its predecessor, is far less expensive, labor-intensive and time-consuming than current mapping technologies. His lab used it to map up to tens of thousands of neurons per animal in a few weeks at a relatively low cost.
The team is now refining BARseq to achieve synaptic resolution for even clearer and more accurate mapping. While BARseq can pinpoint exactly where a neural connection begins, it can only provide a close estimate of where it ends.
“Right now, I can tell you where a neuron sends its axons, but I can’t tell you exactly which neurons it’s forming synapses with,” said Zador. “We still don’t have enough spatial resolution at the destination. But we’re working on it.”
Written by: Charlotte Hu, Content Developer/Communicator | publicaffairs@cshl.edu | 516-367-8455
Funding
This research was funded by the NIH, the Brain Research Foundation, Simons Foundation, the CSHL Cancer Center.
Citation
Chen et. al, “High-throughput mapping of long-range neuronal projection using in situ sequencing” appeared in Cell on October 17, 2019
Principal Investigator

Anthony Zador
Professor
The Alle Davis and Maxine Harrison Professor of Neurosciences
M.D., Ph.D., Yale University, 1994