Cold Spring Harbor, NY — Mutations in the breast cancer susceptibility gene BRCA1 are responsible for approximately 45% of inherited breast cancer and more than 80% of inherited breast and ovarian cancer. Certain “nonsense” mutations in the BRCA1 gene are known to cause RNA splicing defects (“exon skipping”), which lead to the production of abnormal BRCA1 proteins that are missing a particular segment. Until now, scientists have lacked a clear understanding of this phenomenon, which they term nonsense-associated altered splicing, or NAS.
 A recent study by researchers at Cold Spring Harbor Laboratory explains the mechanism of NAS and other related RNA splicing defects. The study has important implications for understanding and treating not only breast and ovarian cancer, but many other diseases including cystic fibrosis, Fanconi anemia, Duchenne muscular dystrophy, and one of the most common inherited disorders in humans, neurofibromatosis.
A recent study by researchers at Cold Spring Harbor Laboratory explains the mechanism of NAS and other related RNA splicing defects. The study has important implications for understanding and treating not only breast and ovarian cancer, but many other diseases including cystic fibrosis, Fanconi anemia, Duchenne muscular dystrophy, and one of the most common inherited disorders in humans, neurofibromatosis.
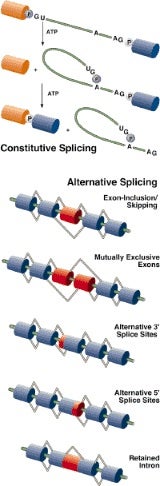
Within genes, DNA serves as a template for the production of messenger RNA, which in turn is a template for the production of proteins. Messenger RNA molecules typically contain protein-coding regions called “exons” as well as non-protein-coding regions called “introns.” Introns are removed from messenger RNA by a splicing mechanism that joins exons together.
The BRCA1 gene encodes 24 exons. Thus, 23 interspersed introns must be precisely snipped away from BRCA1 messenger RNA and the remaining 24 exons must be accurately spliced together—with no exon skipping—if a normal BRCA1 protein is to be produced. The splicing machinery in the cell nucleus makes 46 cuts and 46 pastes (due to a “lariat” intermediate) to generate a single, properly spliced BRCA1 messenger RNA molecule.
The new study by Adrian Krainer and his colleagues at Cold Spring Harbor Laboratory shows that exon skipping due to mutations in BRCA1 and many other genes is frequently, if not always, caused by the disruption of “exonic splicing enhancers,” or ESEs. ESEs are sequences within exons that stimulate messenger RNA splicing. The study is significant because it offers a new explanation for how diverse mutations in several genes lead to RNA splicing defects and in turn, to various diseases.
Krainer’s research had previously identified four distinct families of ESEs, each of which is recognized by a different splicing factor. ESEs and the factors that recognize them probably direct the splicing machinery to the correct sites on messenger RNAs. The new study shows that point mutations which alter the sequence of an ESE can cause exon skipping, most likely by blocking splicing factor binding.
To study the mechanism whereby mutations in the BRCA1 gene cause exon-skipping, Krainer and his colleagues created normal and mutant BRCA1 test genes and mixed them with an enzyme that transcribed the test genes into RNA. Then they added these RNA transcripts to cell extracts capable of splicing RNA.
As expected, the normal BRCA1 test genes gave rise to properly spliced RNA containing the normal arrangement of exons 17, 18, and 19. When the test genes contained the nonsense mutation known to be associated with breast and ovarian cancer, the researchers observed a splicing defect. Namely, exon 18 was skipped, and exon 17 was spliced, abnormally, to exon 19. This was the expected result.
However, the scientists believed that there was nothing special about the particular way that nonsense mutations might disrupt an ESE. They suspected that any kind of mutation that disrupts an ESE—even a “silent” or synonymous mutation in DNA that does not alter protein coding—might cause exon skipping and lead to the production of abnormal proteins. They were correct. When Krainer and his colleagues tested a BRCA1 gene that contained a “missense” mutation, they observed the same splicing defect that they observed with the test gene containing a nonsense mutation.
In collaboration with Krainer, Cold Spring Harbor Laboratory scientist Michael Zhang has developed a computer-based method for predicting the presence of ESEs within genes of particular interest. The scientists used the method in this study to show that the BRCA1 nonsense mutation disrupts one of several predicted ESEs in exon 18. In addition, Krainer and Zhang found that nonsense, missense, and synonymous mutations in several other human disease genes are likely to disrupt ESE function, cause exon skipping, and lead to the production of abnormal proteins that lack a particular segment. These genes include CFTR (cystic fibrosis), DMD (Duchenne muscular dystrophy), FANCC (Fanconi anemia), and NF1 (neurofibromatosis).
When assessed only at the level of a person’s DNA, missense mutations are typically assumed to cause single amino acid changes in proteins, nonsense mutations are typically assumed to create “stop” codons that result in the truncation of proteins, and synonymous mutations are typically assumed to have no effects whatsoever. This study demonstrates that these assumptions are frequently invalid. It highlights the importance of testing whether mutations in coding regions of genes affect RNA splicing when classifying mutations for diagnostic, therapeutic, and other purposes.
In addition, this study suggests that person-to-person variations in ESEs (and other elements that regulate RNA splicing) may account in part for the different effects that some gene mutations have in different people. Variations in ESEs may also account in part for the variable response of patient subpopulations to treatment regimes, and for the variable severity of clinical symptoms. Finally, this study* underscores the notion that the evolution of exon sequences is constrained not only by protein function, but also by the presence of signals critical for proper messenger RNA splicing.
*Liu et al., Nature Genetics, January 2001, pp. 55-58.
Written by: Communications Department | publicaffairs@cshl.edu | 516-367-8455