Imagine a book written with all the right words, but no spaces between words, no punctuation, no chapter headings—it would look like a senseless string of letters one after the other. That is what the first draft of a genome sequence looks like. To make sense of the long string of letters, scientists have programmed computers to break the sequence into chapters, words, and sentences by looking for known patterns. In that process, researchers can quickly identify genes, regulatory elements, and bits and pieces that get clipped out on their way to directing the cell’s protein-making machinery. But computers can get confused by patterns that are not quite what they were programmed to seek. That is where humans come in. People can correct the computer’s errors, making the genome more useful to researchers.
But it doesn’t take a Ph.D. to contribute to the scientific process. High school educators recently joined full time researchers to update the maize (corn) genome at a genome “annotation jamboree”—a sort of DNA interpretation party. CSHL adjunct professor and USDA research scientist Doreen Ware helped organize the event. She admitted that the setting could have been better…
“But that’s because we were supposed to do this in Hawaii,” she laughed.
The 62nd Annual Maize Genetics Meeting was slated to be held in Kailua-Kona, Hawaii, starting March 12, 2020, and the Maize Gene Structure and Function Annotation Jamboree was set to kick things off. However, because of COVID-19 travel restrictions, Ware and jamboree organizers Marcela Tello-Ruiz and Cristina Fernandez-Marco had to rethink how they were going to host their event. They needed to get more than two-dozen people to simultaneously assess genetic databases from their homes. The only way to get together was online.
“Things changed only a few days before we were supposed to board our planes,” Tello-Ruiz said, “but the event was still very successful!”

The computer’s automated review of the genome is just a first draft idea—a model—to describe how an organism uses its genome.
“They’re automated models of what the genes look like, but they’re still models. The genes have not been looked at by humans,” said Ware, who led the creation of the most accurate maize reference genome to date. “One of the things you learn, after you’ve been doing this for a while, is that no matter how good your model is, there are still problems with it.”
That is why the maize annotation jamboree was so helpful. Twenty-eight participants worked together through online video chat sessions to understand what genes were in the genome and what they do. Ware pointed out:
Using visual editing tools like Apollo (named after the Greek god of divination and truth) the jamboree participants reviewed computer-generated annotations that looked suspicious. In some cases, they even validated their corrections with laboratory experiments.
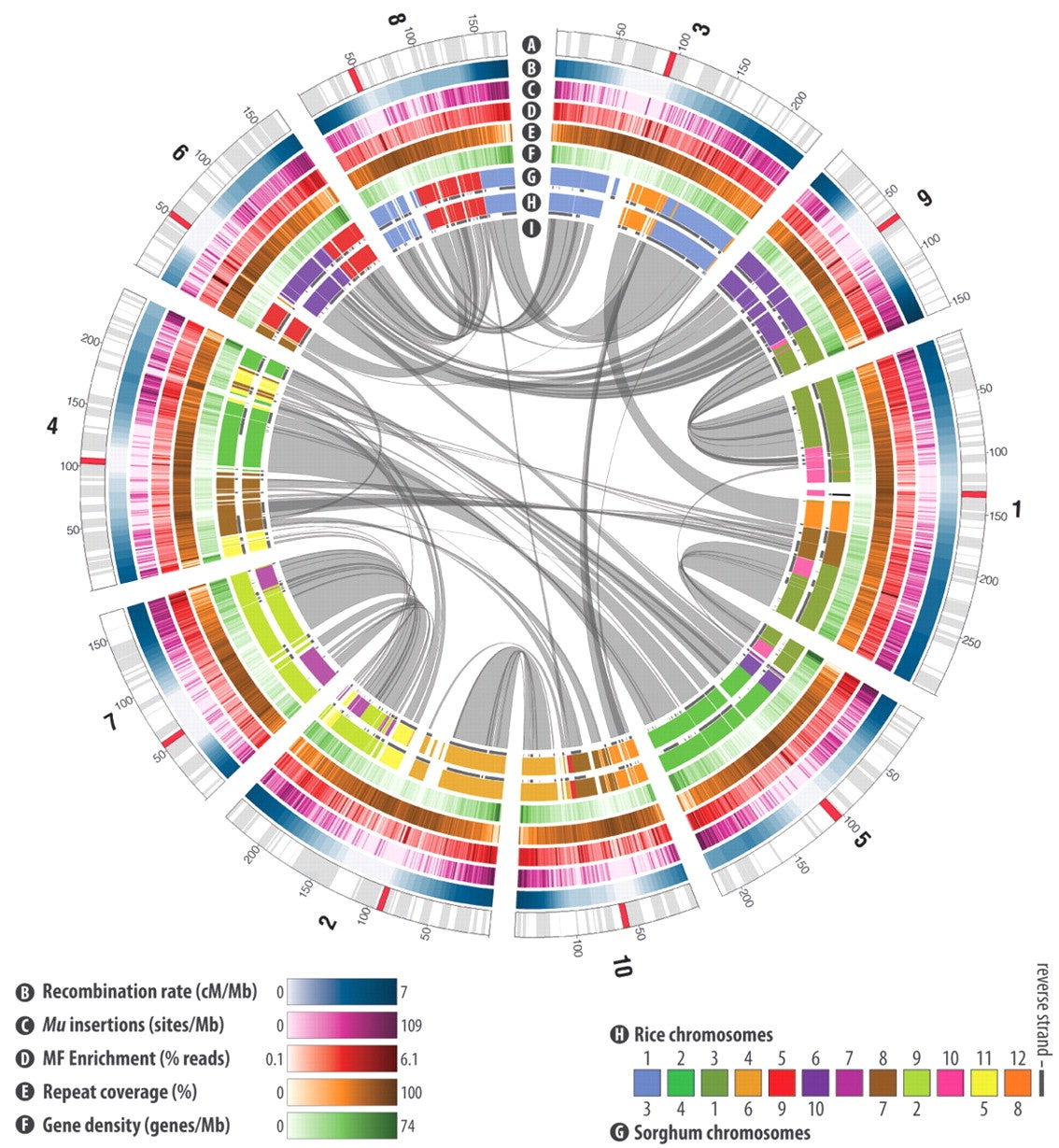
For Ware, this provides a unique opportunity to look for patterns in the errors that computers make. Finding these patterns “paves the way for improving these computational methods,” she said.
And at a time when everyone needs to figure out how to work and learn from their homes, “as we learned with this most recent jamboree… it also could help support education in a time when we’re going to be doing more and more remote work.”
This was the fourth annotation jamboree co-hosted by CSHL’s DNA Learning Center (DNALC) and the first that was done remotely.
“Having students annotate a gene and figure out gene structure—the part of it that carries information and controls how it works—helps them understand what a gene really is,” said Dave Micklos, executive director of CSHL’s DNALC. “Now, through jamborees like this, we can theoretically train people to do this work for whatever genome they might think is fun—the koala, praying mantis, whatever! … This really represents how CSHL is working at the boundary of high-level research and education.”
Fenandez Marco, a DNALC high school educator, added that in the future, they hope to introduce jamborees as a means for scientists to collaborate with classrooms. That way, scientists save time, “and students study what is being researched in a real laboratory.”
- Machines sequence the genome, one letter at a time.
- Computers annotate the genome, mapping out key features like genes and regulatory elements.
- Humans correct, annotate again, and curate the results.
Written by: Brian Stallard, Content Developer/Communicator | publicaffairs@cshl.edu | 516-367-8455
Funding
This work was supported by the National Science Foundation, Gramene, and the USDA Agricultural Research Service.
Citation
Learn about the strategy: Tello-Ruiz and Marco et al, “Double triage to identify poorly annotated genes in maize: The missing link in community curation,” PLOS One, 28 Oct, 2019