Base Pairs podcast
Did you know? If unwound and tied together, the strands of DNA in one cell would stretch past the entire length of you body (~ 6 ft). Now imagine that among all that, only enough genetic information to run the length of your thumb-nail is of any importance!
Soon after the Human Genome Project unveiled the sequence of our DNA, many scientists hypothesized that only a small portion—about two percent—of the human genome actually encodes the proteins that make our bodies work. The rest, they said, was “junk.” This stunning assertion became a widely known bit of trivia—one that led many experts to focus only on what they referred to as “genes,” the protein-coding portion of the genome.
In this new episode of Base Pairs, co-hosts Brian and Andrea talk to David Spector and Thomas Gingeras, two CSHL professors who have realized—in their own unique ways—just how wrong this assumption has been.
AA: And I’m Andrea
BS: And welcome to season two of Base Pairs!
AA: This episode is actually the second half of a two-part season premiere, so if you’re interested in hearing the whole story, we highly recommend you check out our episode from March of 2017. It’s titled Dark Matter of the Genome, Part 1.
BS: And in that episode, we talked to CSHL Assistant Professor Molly Hamell – who coincidently used to be an astrophysicist – about the “Dark Matter” of the genome. The chunk of the genome—
AA: About 98% actually!
BS: —the HUGE chunk that in fact does not code for proteins.
BS: Now before we go any further, I wanted to invite our listeners to join me for a step back into the past – when I was just an awkward high school student trying to pass his biology class.
(classroom chatter)
AA: Oh, my. I’m not sure you’re going to keep an audience for this one, Brian. Are we going to have to hear all about your high school angst?
BS: No… no, no. Nothing like that. But like most of our listeners, I loved science, even then… even if I was sleeping through some of the lectures. And a favorite wacky science fact my teachers – and even the textbooks! – tended to repeat was that more than 90% of the genome was… junk. “Junk DNA,” is what everyone called it. And it stuck. I remembered that. Even when I turned my attention away from biology to space science, and animal science, and climate change, and medical reporting… that crazy statistic stuck with me.
AA: Well, that’s why we prefer the term “Genomic dark matter” so much more. Instead of seeing “junk,” the world is finally learning to look the non-coding part of the genome – the part that doesn’t contain the recipe for proteins — as the exciting unknown, an area worth investigating.
BS: Right, right… but imagine being a scientist interested in that unknown back when I was in high school. When everyone, even your esteemed colleagues, also pushed the “junk DNA” concept. At the time, people researching that ignored part of the genome must have looked a little like… well… garbage pickers, or maybe trivia freaks.
TG: There was sort of a negative connotation associated with any region of the genome that was not engaged in making proteins. And, it acquired a label of Junk DNA, in the literature, and therefore, this pejorative concept really stayed with us and focused the attention of most people on these protein coding regions, without the need for studying these other regions.
BS: That’s Tom Gingeras, a CSHL professor who can be found carefully scrutinizing genomes at the aptly named Genome Research Center.
TG: Even when I was an undergraduate, one of the major topics that was being pursued by a lot of different labs, and this will sound somewhat strange given where we are today, was that where were genes in DNA? Obviously, we had an understanding that DNA was the genetic molecule that’s passed from one cell type to another, and from one generation to another. But we didn’t realize that a unit called a gene … What was it? How was its structure identifiable in gene, in a genome? How was its expression, how was its information turned on at any one particular time, in one cell type versus another? These are questions that continue to be pursued in science. But, in the days when I was a student, we knew virtually nothing about it. That’s been sort of a probing question for me for many, many years.
AA: Oh wow. I can see why he’d want to be a part of that. It’s the one drive A LOT of scientists share: a desire to break new ground in the search for information about… everything!
BS: That’s it, and to make those new discoveries, Tom found himself leading projects were he and his lab wadded and sifted through the genome’s so-called junk, one nucleotide at a time. It was all part of the ENCODE project-
AA: That’s E-N-C-O-D-E – an international project consortium first established by the U.S. government in 2003 to examine the recently decoded human genome sequence in greater depth.
BS: Yup. And they were spending most of their time studying the areas occupied by what’s considered “genes” – that 2% of the genome that leads the production of proteins.
TG: So, we… looked at two chromosomes. Chromosomes 21 and 22. And we walked along the genome roughly every five bases. Is there an RNA that contains that base as encoded in the genome? And then you’d walked to the next five and ask, “Is there an RNA coming from there?”
AA: To remind our listeners: RNA is the short-lived cousin of DNA. Scientists first learned about RNA because it carries the instructions for making proteins from the DNA to the cell’s protein factories.
BS: Right, and with that understanding, most scientists looked for the presence of functioning RNA as a sign of activity coming from the protein-coding parts of a genome. In this way, many hoped to figure out where exactly each gene is on a chromosome, and what it’s up to. Tom figured that since only 2 percent of the genome was genes, only about 2 percent of each chromosome would be written in an RNA—or transcribed, as scientists say.
TG: My post-doc at the time, Phil Kapranov, came back with a result, which seemed to me entirely wrong because it went against the very simple principle that the only place that we probably should be seeing things are where known genes are… Now, you might see new known genes, and so, all right, so you would increase the number of spots that would be functional in that fashion. But, instead, we saw almost a totality of those two chromosomes being transcribed. So, I got very upset with Phil, because he obviously did the wrong experiment and what he did was wrong. I asked him, “Please, go back and do this again.”
BS: So. Phil did it again, and again, and again – each time with the same result.
TG: The exact same result…
AA: Ok. So NOW I’ve got to know. What’s going on here?
BS: You know? That’s the thing. Tom had no idea. As far as the scientific community was concerned, Tom’s lab had been sifting through junk, looking for treasures. But now it seemed that EVERYTHING was treasure! The genome in those 2 chromosomes was expressing massive numbers of RNA messages, or transcripts, as they are called by scientists.
TG: To add to the complexity… this incredibly rich output of RNA from every region of the genome indicated that there was a lot of energy being “spent” by cells being put into making these RNAs, which apparently had no function since many of them– most of them– were well outside the 2% of the genome that encodes protein.
AA: To blow so much energy on making useless RNA… that doesn’t sound like the efficiency we’ve come to associate with cells. Nature, as many experts will tell you, generally has a “waste not” kind of attitude.
BS: Exactly. After seeing examples of this phenomena again and again, Tom and his team decided to change how they perceived the genome. Almost overnight, they became those so-called “garbage picking” scientists I mentioned earlier. And in doing so, they discovered something that really abolishes the “junk DNA” mythos.
TG: What we determined is that roughly 80% of the genome is transcribed… Not every cell makes 80% of its genome, but if you look at a totality of many, many different cell types, what is capable of being transcribed is about 80% of the genome. Only 2% of the genome is actually embedded within a protein coding region. So, that meant that a majority of the genome was transcribed and that most of it was making RNAs that were not intended to make a protein.
AA: 80%… Wow. So… we know today that there’s around 20,000 protein-coding gene regions. Scientists even today are still trying to identify what each gene does. Tom’s work is exciting stuff – don’t get me wrong – but can’t we just… I don’t know… shelve it until we get the protein coding parts figured out?
BS: (laughs) Well…Initially, that’s exactly what happened. A lot of scientists simply didn’t accept Tom’s results, saying that Tom, Phil, and rest must have been mistaken.
TG: ‘…either your technology doesn’t work, or B, it doesn’t make any difference because it’s all noise anyway because it’s not a perfect system.’
BS: And then, even among the people who did believe it, they argued that – like you said – it’s just too much. They preferred to focus on what the Human Genome Project had made known, and suggested saving the unknown for a rainy day.
AA: But then… something happened, didn’t it?
TG: Slowly but surely, what we saw was that these RNAs were seen to be important regions, that if they were mutated, deleted, you would see an effect. A phenotypic effect.
BS: By “phenotypic effect,” Tom means, simply, a physical manifestation in living creatures – things that made them look different, or in some cases, made them sick. Our first sense of the scope of these changes came about 5 years ago, when teams from 32 institutes in five countries – all contributing to the ENCODE project – released 30 papers at once.
AA: Not exactly light reading.
BS: Hehe. Well, it was important stuff! Previously, it had always been assumed the only way to affect the physical expression of traits was to mess with a gene, or – as we described in the previous episode – inhibit the regulation of transposable elements. But a lot of the ENCODE data showed that even if you messed with only the non-protein coding parts of the genome, it could affect cells, and whole organisms, in a big way.
AA: Oh wow… that is important. I take it the scientific community is a bit more amenable to Tom’s results now, right?
BS: Well…
TG: I’ll answer that with a personal vignette. So, I would go to meetings and we would present our data. That almost invariably ignited a discussion, no matter how large the audience was. I’ve been in places where there’ve been more than a thousand people, and then people would have no problem jumping up and saying, “This is all nonsense. And even if it is all true, we are totally swamped with just learning about the things we know about, instead of dealing with all this other nonsense.” You would have this back and forth. I would go back home feeling very upset. Feeling, “why can’t they see? Why can’t they understand?” And I would say, “We’re not incompetent. So why is that we’re having this discussion?” And I would get very personally worked up. Until one day I honestly had what might be called, in sort of a religious sense, this enlightenment. Seriously. Sitting in my office. Which, basically, led me to understand that you need to let them discover it…
I’m sure that people will do these experiments in their own way in their own systems. And, if they see the same thing then perhaps you’ll understand that there’s a lot of work to be done. And, that the things we thought were solved, in fact, are not solved, and they offer an opportunity to learn even more.
DS: … Yes, 7SK. Yeah. That kind of started our interest and then it just expanded from there. Now it’s pretty much the main focus of my entire lab. We do very little imaging at this point. Mostly we’re doing RNAseq and try to figure out the function of these long non-coding RNA’s.
AA: That’s Professor David Spector – and he’ll explain “7SK” in a minute….
BS: David was once one of “them.” Those doubting scientists Tom was talking about. Not necessarily a critic of ENCODE’s work. Just a scientist so focused on his own work that he didn’t pay much attention to the non-coding part of the genome.
AA: You see, David is the Director of Research here at CSHL, so we know a lot about him…
BS: And we know he’s REALLY adamant about speckles.
(Clip from the holiday party) (laughter fades)
DS: My lab is probably best known for the early work that we did on nuclear organization and function. I started that work probably back in 1981. We were very intrigued by how the nucleus might be organized, given the fact there are no membrane-bound structures in the nucleus. Yet if you stain cells with either dyes or with antibodies there are clearly concentrations of specific proteins in different places in the nucleus… So, I got really interested in studying these structures and spent a significant amount of my scientific research career on a particular nuclear structure called nuclear speckles…
…in fact, some people call them Spector speckles as a joke. We spent a lot of time working on them. We purified them biochemically. Did proteomic analysis, identified 146 proteins in them. We did live cell imaging of them and just an enormous amount of work.
BS: So much work. Back then, the majority of David’s lab was dedicated to this subject, but that started to change when he had his own run-in with the unknown.
DS: “7SK. It was a known RNA at the time”
BS: Ok. So not THAT unknown. But it was still a non-coding RNA, and the fact that it was jumbled in there – hiding in Spector’s speckles – that caught his attention.
DS: that kind of tipped the bucket in a way from proteins to RNA because we were intrigued by the fact that there was such a high concentration of this non-coding RNA in nuclear speckles.
AA: I see. Here’s this important region of the cell, and David found himself wondering, “why is it wasting energy making so much of this supposedly useless RNA?”
BS: That’s the ticket. But David’s foray into the Dark Matter of the genome didn’t stop with just one RNA.
DS: One day we got a call in the lab from a student in a lab in France
BS: — her name is Delphine Bernard —
DS: She was doing a study in neurons and… She came upon a long non-coding RNA and her adviser was telling her to drop it and forget about it and focus on what his lab was interested in. She persisted and she was curious about it and because my lab had been working on long non-coding RNA’s she decided to call us and see what do we think? So, she started to tell me about this RNA on the phone and it sounded really exciting so we told her absolutely don’t drop it. We started a collaboration with her and then we ended up having a paper together in EMBO Journal. Her PI bought into it very strongly, so he was a convert. (laughter) Anyway, so that kind of got us interested. The RNA happened to be MALAT1.
AA: Ok. That sounds VERY familiar. MALAT 1 is a puzzle, even among genomic dark matters.
DS: MALAT 1 in essence breaks the rules for all long non-coating RNA’s. First rule, most long non-coding RNA’s are present in very few copies per cell. MALAT 1 is widely abundant in cells. In fact, in many cell culture lines it rivals… one of the most highly expressed transcript in tissue culture cells
AA: And that… that would indicate that MALAT 1 is important.
DS: It’s got to have a really amazing function.
AA: VERY important.
BS: And yet, when they studied mice that couldn’t produce MALAT 1…
DS: At the end of the day, the mouse was perfectly happy without MALAT 1.
AA: What?!
BS: Yeah. Perfectly healthy. David’s lab even sent a number of tissue samples to folks over at Yale.
DS: They looked at all the tissues, couldn’t find anything abnormal about them. The mice have been in my lab now breeding perfectly well for probably close to seven years.
AA: Somehow, I doubt this was a complete bust. It seems highly unlikely that cells mass-produce MALAT1 for fun.
BS: that’s what David thought too. He figured that MALAT1’s function may in-fact be more important than he and his lab presumed – SO important that healthy cells have back-up systems in place if the RNA’s production fails in one part of the genome.
DS: Given that, we said “okay we need to take a different approach…”
AA: So, David and his team needed to study cells that were omnivorous — hungry for all things good for them… (Pause) that sounds like…
BS: Yup. Cancer. Aggressive tumor cells are like regular cells gone rogue, multiplying rapidly and sapping up important resources in the otherwise organized world of a living thing. Tumor cells that travel from one place to another – such as breast cancers that invade the lung – are called metastatic – and you can often blame their bad behavior, at least in part, on a mangled genome.
AA: So mangled that the failsafe in place of MALAT1 might not work. I see. Take MALAT1 away from a tumor, and something might happen.
BS: Something dramatic.
CBSNY: “A possible breakthrough tonight in the war on Breast cancer…” CG: “It was a eureka moment when a new drug he and colleagues invented, chewed up and destroyed aggressive metastic breast cancer cells.” DS: “The effect was so dramatic that it was not something I could have actually predicted…”
AA: That was David talking to Carolyn Gusoff of CBS2 New York just last year – and his words… they’re not the kind that scientists say lightly. The effect REALLY was dramatic. So much so, that David has microscopy photos of the experiment framed on his desk.
(in interview) AA: so, I’m looking at this picture that’s right behind you. So that kind of captures the difference that you saw between– So can you describe that a little bit, like what it looks like? What you saw that was so different about the chamber without MALAT 1?
DS: When we looked at the tumors in this mouse model that had MALAT 1, the tumors were very aggressive tumors which means that the tumor was just filled with cancer cells.
BS: I think the best way to describe this is to imagine a white sponge, except… there are LOTS of wide holes in this sponge and each of them is PACKED full of this pink… stuff.
AA: Those are the cancer cells. The pink stuff.
DS: When we got rid of MALAT 1, the tumor totally changed. A lot of the cancer cells died and were released and the tumor changed its whole gene expression pattern and it started to form these cystic structures which were formed because cells died, leaving these cysts.
BS: Interestingly, these large cavities, once rife with aggressive pink cancer cells, they’re now full of something much more benign: Milk.
DS: these cysts are filled with liquid – milk proteins and so what does that mean? If you think about it, this is a tumor in a mammary gland and at a particular stage a mammary gland can produce milk. What we’ve done is by taking away MALAT 1 we’ve changed that tumor from being this aggressive tumor, to …
AA: To one that has switched its focus to milk production. You can see these images for yourself at our LabDish blog to really feel that “wow factor” for yourself. And David and his colleague think it’s safe to assume that if they turn off MALAT1 in tumors in other parts of the body, the cancer cells will be replaced with other types on harmless proteins.
BS: The coolest thing about all of this is that MALAT1 is just one of over 17,000 known non-coding RNAs. Few of these are as prevalent as MALAT1, and even fewer are seemingly as crucial, but there is an awful lot about non-coding RNAs that we still don’t know.
AA: And that 17,000 is just the ones recorded so far. (pause) According to David, by targeting MALAT1 and parts of the genome like it, experts might be able to open up new avenues for personalized treatment options in cancer and possibly other illnesses.
DS: The long-term goal of this over the next ten years, let’s say, is to develop a precision medicine based approach whereby we could get a small piece of the patient’s tumor. We could put it through a battery of tests… and screen them for these non-coding RNAs and identify which patients would benefit most from knocking down these three RNAs versus another three. That’s kind of what we hope to do because every patient’s tumor will be different.
BS: Since David’s work has taken off, MALAT1 has really become a poster child for the importance of what was once thought of as “junk DNA.” And according to Tom Gingeras, the opinion of the scientific community is changing too.
TG: Most of the scientific community, at least I come in contact with, have an appreciation for the fact that these regions we thought were relatively inactive, that is to say, nonfunctional, probably contain some proportion of things which are of biological note. There’s an argument about how much that is, and then there’s an argument exactly how important. But, there is, I think, a general appreciation that it’s not something we just slide under the carpet.
AA: So… that’s it! While research into the non-coding part of the genome is just getting started, this is where our story is going to have to stop.
Extras for Episode 9
Explaining ENCODE
Professor Thomas Gingeras is not only a leader of the ENCODE project, but also the mouseENCODE and modENCODE (model genome ENCODE) projects of the National Institutes of Health. His research has altered our understanding of the traditional boundaries of genes, but what is an ENCODE project all about anyway? Below is a pair of videos, produced by Nature in 2012, that help explain:
Ten years on from the Human Genome Project’s completion, “the next colossal chapter in your story” begins: an ENCyclopedia of DNA Element, or ENCODE for short.
ENCODE’s lead coordinator, Ewan Birney, and Nature editor Magdalena Skipper talk about the challenges of managing this colossal project and what we’ve learnt about our genomes.
You can view all the ENCODE results, including the 30+ papers we mention in this episode by visiting Nature’s ENCODE Explorer tool.
Seeing white
And remember how we promised we’d show you that photo of Dave’s breathtaking results from when he investigated Malat1? Well, here they are:
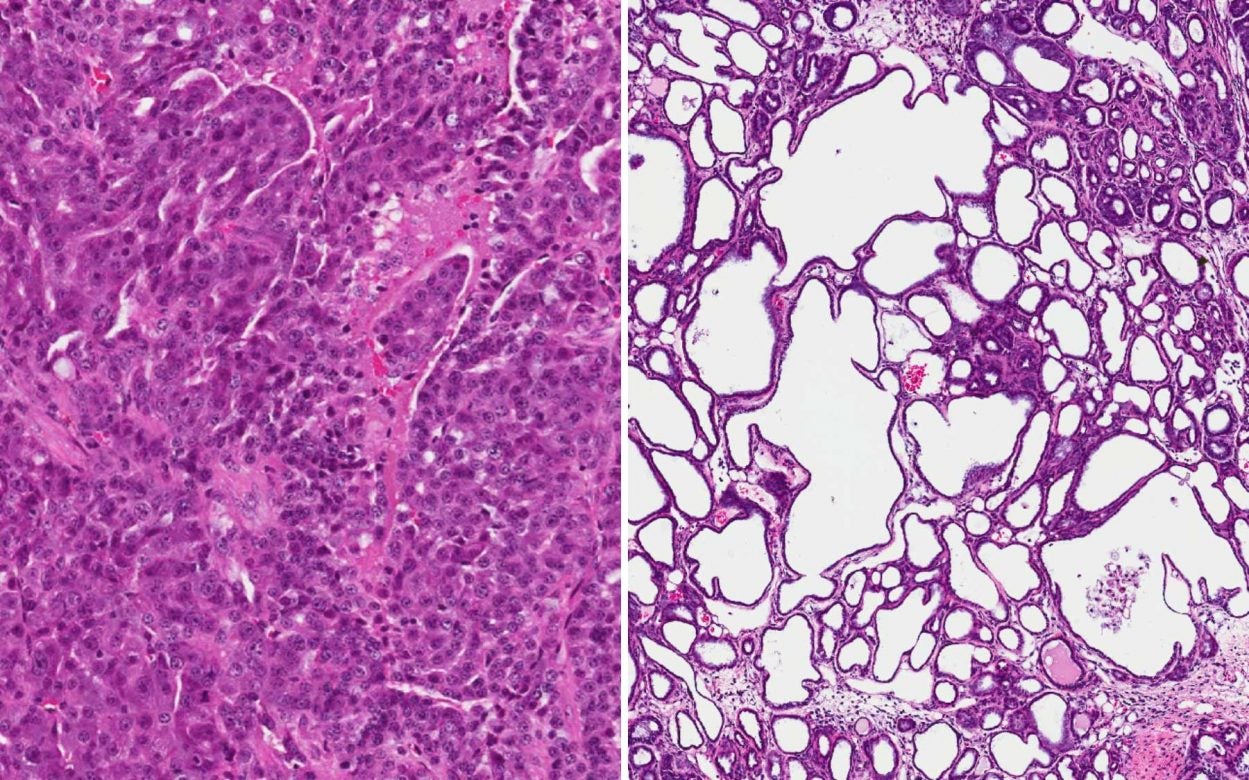
You can learn more about these incredible results at: https://www.cshl.edu/unusual-drug-target-and-drug-generate-exciting-preclinical-results-in-mouse-models-of-metastatic-breast-cancer/
“Differentiation of Mammary Tumors and Reduction in Metastasis Upon Malat1 LncRNA Loss” appeared online in Genes & Development on December 23, 2015. It was authored by Gayatri Arun, Sarah Diermeier, Martin Akerman, Kung-Chi Chang, J. Erby Wilkinson, Stephen Hearn, Youngsoo Kim, A. Robert MacLeod, Adrian R. Krainer, Larry Norton, Edi Brogi, Mikala Egeblad and David L Spector.
Written by: Brian Stallard, Content Developer/Communicator | publicaffairs@cshl.edu | 516-367-8455
