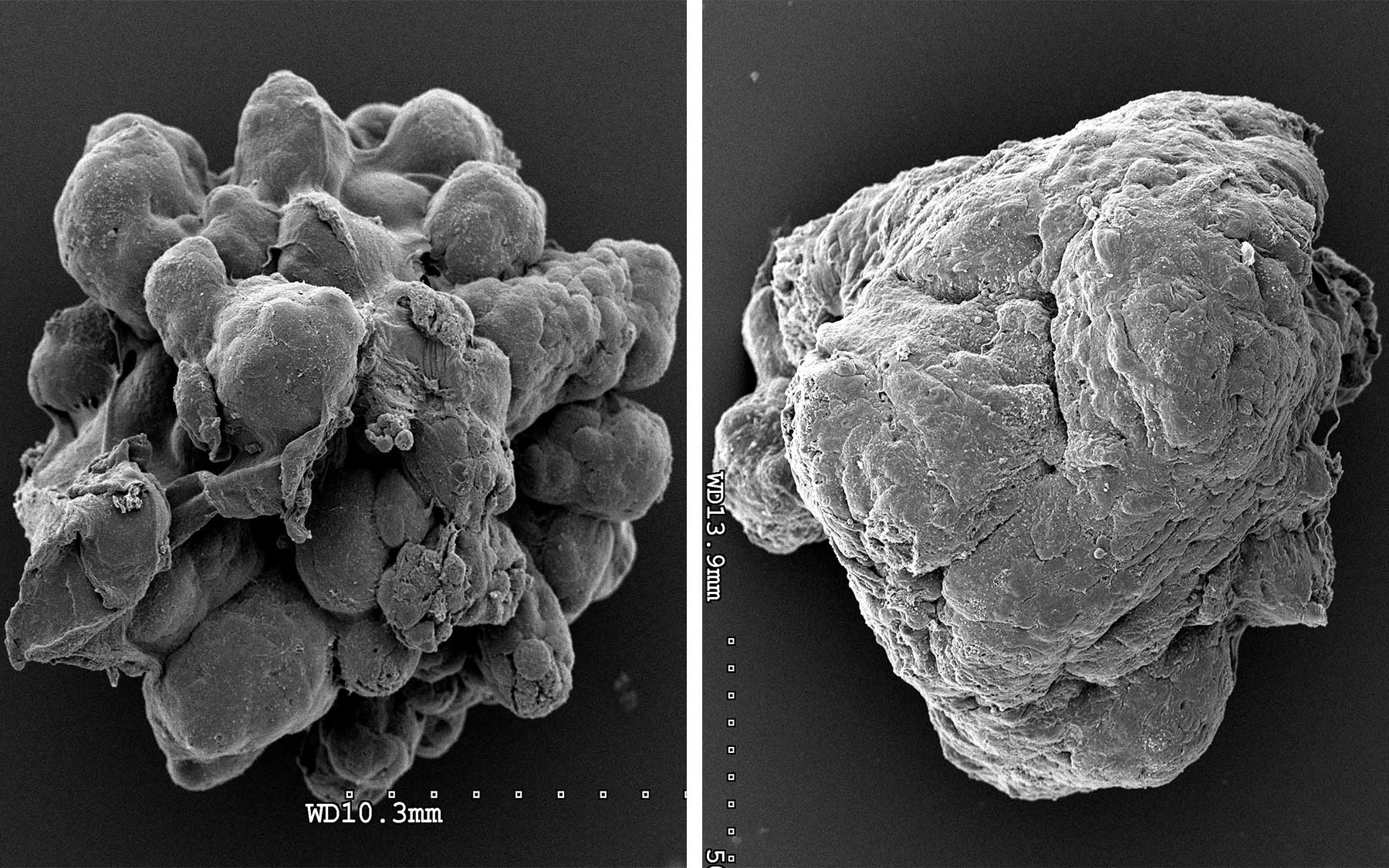
CSHL scientists test an antisense method of targeting long noncoding RNAs overexpressed in breast cancers
Cold Spring Harbor, NY — The human body produces 100,000 or more different proteins. Yet, amazingly, only two percent of the human genome actually encodes proteins. Nearly 80 percent of the rest of the genome is transcribed into RNA that does not code for proteins. Two big questions facing scientists are: How much of this “non-coding” RNA is actually functional? And does it play a role in disease?
A team of scientists at Cold Spring Harbor Laboratory (CSHL) screened thousands of non-coding RNAs to find those that were expressed at high levels in two types of aggressive breast cancer. As they describe today in a paper appearing in Cell Reports, when they reduced the level of some of the most over-expressed of these RNAs from mammary tumor samples, cellular features characteristic of cancer spread were significantly reduced.
Of the handful of different types of non-coding RNA, the most abundant and least understood are long non-coding RNAs, or lncRNAs. About 16,000 lncRNAs have been identified in humans, but functions for the vast majority are unknown.
“Since so much of the genome is being transcribed into RNA, it would seem that there would be a vast wealth of potential therapeutic targets out there that have not really been studied,” says the team leader, CSHL Professor David Spector, who is also Director of Research at the Laboratory.
While the exact functions of most lncRNAs remain to be discovered, it has already been shown that in some cases their over-expression is linked to specific cancers, including breast cancer, prostate cancer and leukemia. Earlier this year, Spector’s team demonstrated that a lncRNA called Malat1 was a critical regulator of breast cancer progression. Eliminating that particular lncRNA in a mouse model of luminal B breast cancer caused the cells within the primary tumor to change character, and resulted in a significant reduction in metastasis.
“That study provided significant motivation for us to look for other lncRNAs that might also be over-expressed and impact breast cancer,” says Spector.
Spector and his team, led by postdoctoral fellow Sarah Diermeier, systematically sifted through the vast database of lncRNAs to identify those that are expressed more often in tumors, relative to normal mammary cells.
The team found several hundred lncRNAs that were expressed at higher than normal levels in both types of aggressive mouse tumors that they tested: luminal B and Her-2 positive. They then performed an extensive computational analysis to prioritize a subset of 30 of these lncRNAs that they dubbed Mammary Tumor Associated RNAs, or MaTARs.
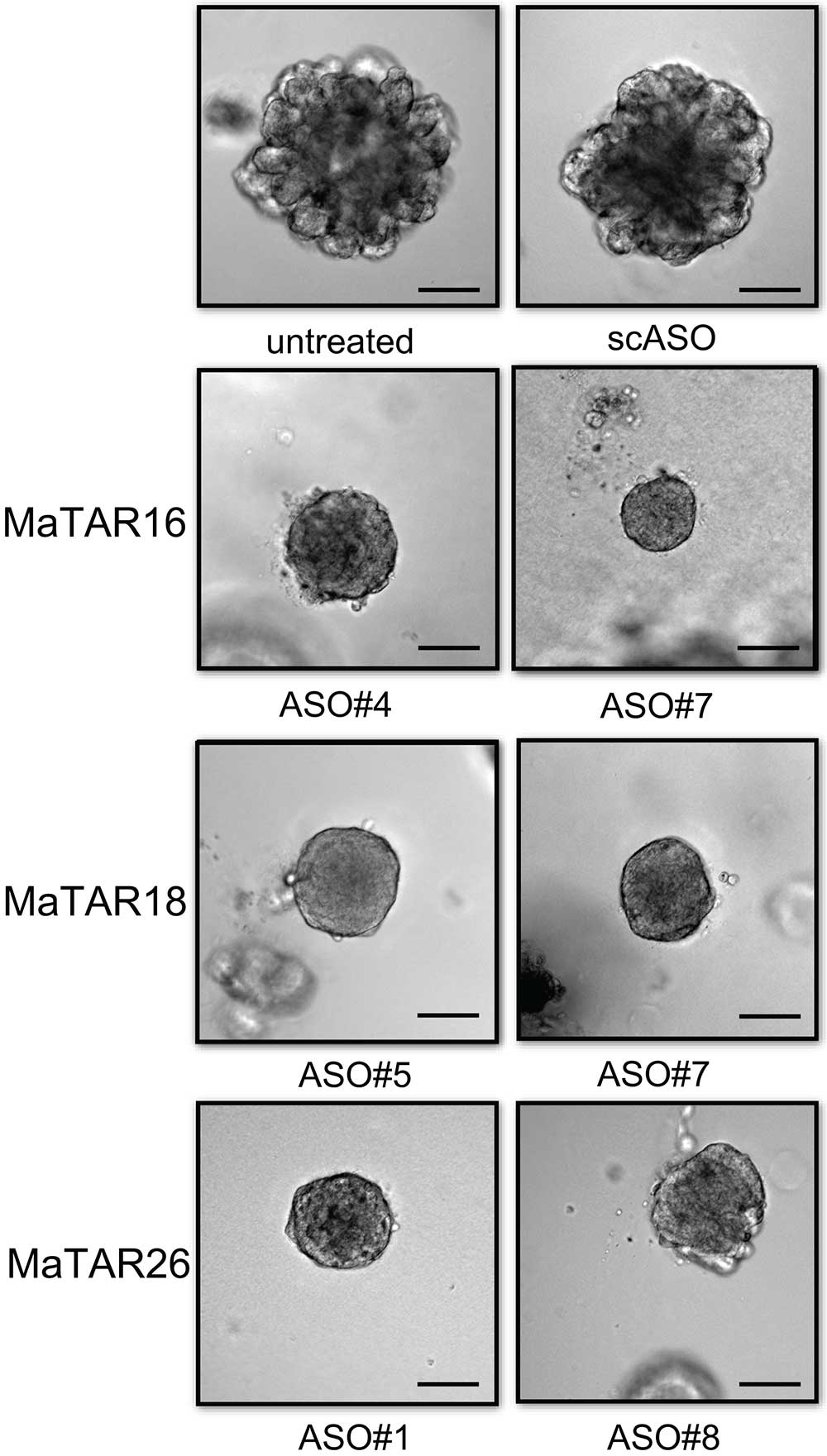
In collaboration with Ionis Pharmaceuticals, Spector and his colleagues designed a series of molecules that bind tightly to, and thereby destroy, specific RNA sequences. They used these so-called “antisense” molecules to wipe out individual MaTARs in mammary cancer-derived organoids, three-dimensional models of tumor cells that represent many features of real tumors.
The researchers found that individually eliminating 20 of the 30 MaTARs in these organoids diminished features associated with cancer, including cell proliferation, invasion, and migration.
“We now have an innovative way of destroying RNA targets inside live cells and assessing whether a tumor is dependent on them for survival,” says Spector.
The team’s next step is administering antisense molecules to degrade specific MaTARs in mice, in the hope that this will decrease primary tumor mass and/or metastasis. Should those experiments be successful, Spector’s team will perform additional preclinical tests in human tumor samples to better identify which subgroups of patients would benefit most from being treated with antisense molecules to eradicate certain lncRNAs or clusters of lncRNAs.
“We think these tests will have particular relevance for personalized medicine,” says study first author Diermeier “We imagine a situation where organoids can be derived from an individual’s tumor, grown up in a dish, and act as a platform for figuring out which antisense molecules comprise the optimal treatment for a patient.”
Written by: Chris Palmer, Science Writer | pubaff@cshl.edu | 516.367.8455
Funding
The research described in this release was supported by a National Cancer Institute grant 5PO1CA013106-Project 3, the Manhasset Women’s Coalition Against Breast Cancer, the Simons Foundation, and a Cancer Center Support Grant to Cold Spring Harbor Laboratory (2P30CA45508).
Citation
“Mammary Tumor-Associated RNAs Impact Tumor Cell Proliferation, Invasion and Migration,” appears online September 27, 2016 in Cell Reports. The authors are Sarah D. Diermeier, Kung-Chi Chang, Susan M. Freier, Junyan Song, Osama El Demerdash, Alexander Krasnitz, Frank Rigo, C. Frank Bennet, and David L. Spector. The paper can be viewed at http://www.cell.com/cell-reports/newarticles
Principal Investigator

David L. Spector
Professor
Robert B. Gardner Jr., Professor
Cancer Center Member
Ph.D., Rutgers University, 1980