Wild tomato genome, sequenced at CSHL, provides basis for understanding how the domesticated variety evolved and adapted to new environments—knowledge that can help improve worldwide tomato production
Cold Spring Harbor, NY — A team of scientists from Cold Spring Harbor Laboratory (CSHL) and their colleagues in the Tomato Genomics Consortium (TGC) have succeeded in a joint effort to sequence the genomes of the domesticated tomato and its wild ancestor. This achievement is expected to greatly impact the billion-dollar tomato industry by speeding up efforts to breed varieties that can better withstand pests, droughts and diseases.
The sequencing of the domesticated “Heinz” or cultivated tomato (Solanum lycopersicum) was a joint effort by the TGC, a group of about 300 scientists from more than a dozen countries. The sequencing of the tomato’s wild ancestor, Solanum pimpinellifolium, was carried out in parallel at CSHL led by Assistant Professor Zachary Lippman, Ph.D., in collaboration with Professor Richard McCombie, Ph.D., of CSHL and Doreen Ware, Ph.D., of the U.S. Department of Agriculture and a CSHL Adjunct Associate Professor. Both sequences and their comparison are reported together in a paper that appears in Nature on May 31. Also available online are genetic and physical maps of the genomes.
Together, the sequences provide the most detailed look yet at the functional portions of the tomato genome, revealing the order, orientation, types and relative positions of approximately 35,000 genes spread across 12 chromosomes. The sequences will help researchers uncover the relationships between tomato genes and traits and broaden understanding of how genetics and environmental factors interact to determine a field crop’s health and viability.
The sequences also offer insight into how the tomato genome has changed over millions of years. The scientists report that it expanded abruptly about 60 million years ago, at a time close to one of the large mass extinctions. Over time, most of this new genetic material was lost, but some genes survived and contributed to the genetic raw material that gave tomato fruits some of their most appealing traits such as taste, texture and size.
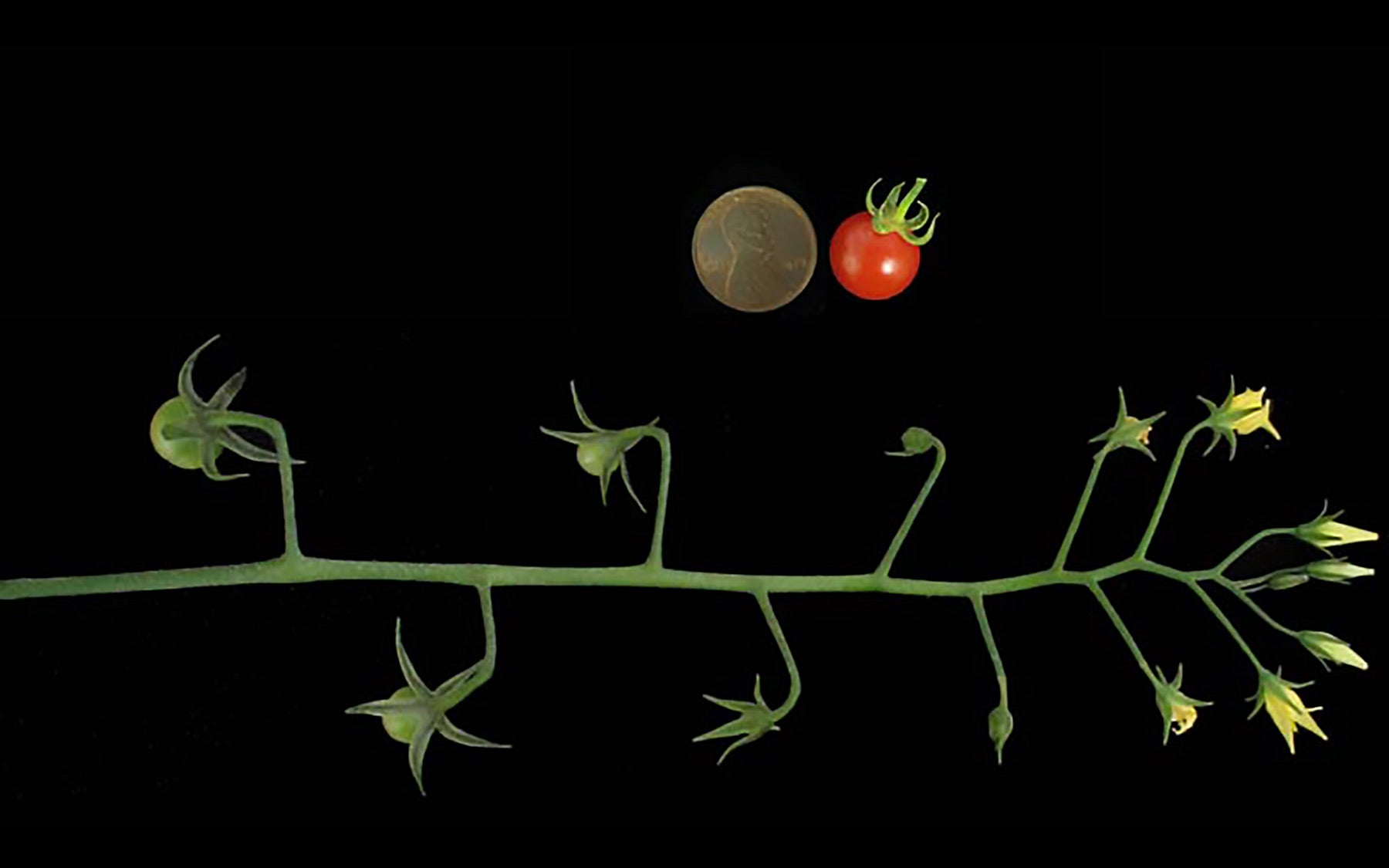
“Comparing the wild tomato genome with the Heinz genome allowed us to explore how the domesticated variety has changed since our ancestors began to select and breed these plants thousands of years ago,” says Lippman. “We were surprised to find that there was a remarkably high degree of similarity between the two genomes.” In another interesting finding, the scientists observed the reintroduction of many large chunks of DNA from the wild species into the genome of the domesticated tomato. These chunks are likely to have brought in disease resistance genes that abound in wild tomato species.
“One key focus for the tomato research community in the future will be to see how many genetic changes it took to go from the small, 1 gram cherry type of tomato that is produced by the wild ancestor to the extremely large domesticated fruit that can range from 30 grams to as much as 2 kilograms,” Lippman explains. “The goal of our group at CSHL is to understand why tomato species vary in the number of flowers they produce. Our sequencing of the Solanum pimpinellifolium genome will certainly help launch this effort, but future sequencing and genetic mapping of other wild species and domesticated varieties will be critical for pinpointing these changes.”
In addition to helping to pinpoint tomato genes with evolutionary and agricultural importance, the two new sequences are also expected to help find important genes in tomato’s relatives within the Solanaceae family, including potato, pepper, petunia and eggplant.
The genome sequence and related resources can be accessed at the Solgenomics website.
Written by: Hema Bashyam, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Citation
“The tomato genome sequence provides insights into fleshy fruit evolution” appears in Nature on May 31 and is embargoed until 1pm EST on May 30. The paper was authored by the Tomato Genome Consortium, which includes more than 300 scientists from a dozen countries including Argentina, Belgium, China, France, Germany, India, Israel, Italy, Japan, Korea, Netherlands, United Kingdom, Spain and the United States, can be downloaded at www.nature.com using the doi:10.1038/nature11119.