Molecular “Handshake” of Key Parasite Protein Revealed as Target for Drug Design and Vaccine Development
By determining the molecular structure of a protein that enables malaria parasites to invade red blood cells, researchers have uncovered valuable clues for rational antimalarial drug design and vaccine development. The findings are reported in the July 29 issue of the journal Cell.
Malaria causes approximately 400 million clinical cases and 2 million deaths annually, with more than 80% of deaths occurring among children. The disease is caused by mosquito-borne parasites of the genus Plasmodium (primarily Plasmodium falciparum). Following the initial stages of infection, merozoite-stage parasites (“merozoites”) invade red blood cells, leading to clinical symptoms and in many cases, death.
“Niraj Tolia [the first author of the study] had malaria when he was young. So when he joined my lab as a graduate student, it didn’t take long for him to convince me that he should be the one to work on this project,” says structural biologist Leemor Joshua-Tor of Cold Spring Harbor Laboratory, who led the research.
A major pathway through which malaria parasites invade red blood cells is the binding of a protein on the surface of merozoites called EBA-175 to a receptor protein on the surface of red blood cells called glycophorin A. Merozoites die if they do not invade red blood cells soon after their release (from liver cells) into the bloodstream. Thus, the binding of EBA-175 to glycophorin A is a prominent target for the development of therapies to control malaria.
To explore the molecular basis of the binding of EBA-175 to glycophorin A—with the rationale that such information might reveal strategies for preventing and treating malaria—the researchers used x-ray crystallography to determine the atomic structure of a key portion of the EBA-175 protein called the RII domain.
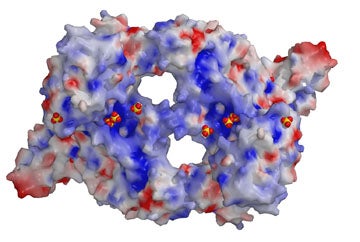
The results revealed that two molecules of RII come together in a manner resembling a handshake, and that the overall shape of such RII “dimers” resembles a donut with two holes (see image above).
Next, to identify precisely which parts of the RII surface bind to glycophorin A, the researchers determined the atomic structure of RII crystallized along with sugar molecules called glycans. Previous work by a co-author of the study, Kim Lee Sim of Protein Potential LLC, established that glycans displayed on the glycophorin A receptor are required for RII binding and for the invasion of red blood cells by the malaria parasite.

The new results showed that each RII dimer binds six glycans. Interestingly, these glycans were discovered to be sandwiched between surfaces where the two RII molecules bind to each other when they form their handshake. This finding suggested that the RII handshake interaction serves to clamp the parasite protein onto the glycophorin A receptor of red blood cells (see image below). An important idea stemming from this view is that blocking the RII interaction—with drugs or vaccines—should block glycophorin A receptor binding and forestall malaria infection.
To test this idea, the researchers created altered versions of the RII protein that they predicted would block the RII handshake, glycan binding, or both. The result: All such altered versions of the RII protein failed to bind to red blood cells, confirming the idea that drugs or vaccines that block the RII interaction, glycan binding, or both might be effective therapies for malaria.
“We now see precisely how a key part of a malaria parasite protein works. This enables researchers to design very specific wrenches to throw into the works. The EBA-175 protein and others related to it appear to be unique to Plasmodium, so they are excellent drug and vaccine targets,” says Joshua-Tor.
Joshua-Tor, Tolia, and Sim were joined in the study by Eric Enemark of Cold Spring Harbor Laboratory.
Written by: Communications Department | publicaffairs@cshl.edu | 516-367-8455