TUTase enzymes interact with let-7, a regulator of development; targeting them could help control certain cancers
Cold Spring Harbor, NY — Over millions of years of evolution, cells have developed myriad ways of regulating the processes that enable them to thrive. Especially useful tools have been saved, or “conserved,” over the eons, so that today we can find them in a broad spectrum of life forms, from the very primitive to the most complex.
Research published July 3, 2017 by structural biologists at Cold Spring Harbor Laboratory (CSHL) sheds light on how one such conserved mechanism works, with implications for the development of new cancer drugs.
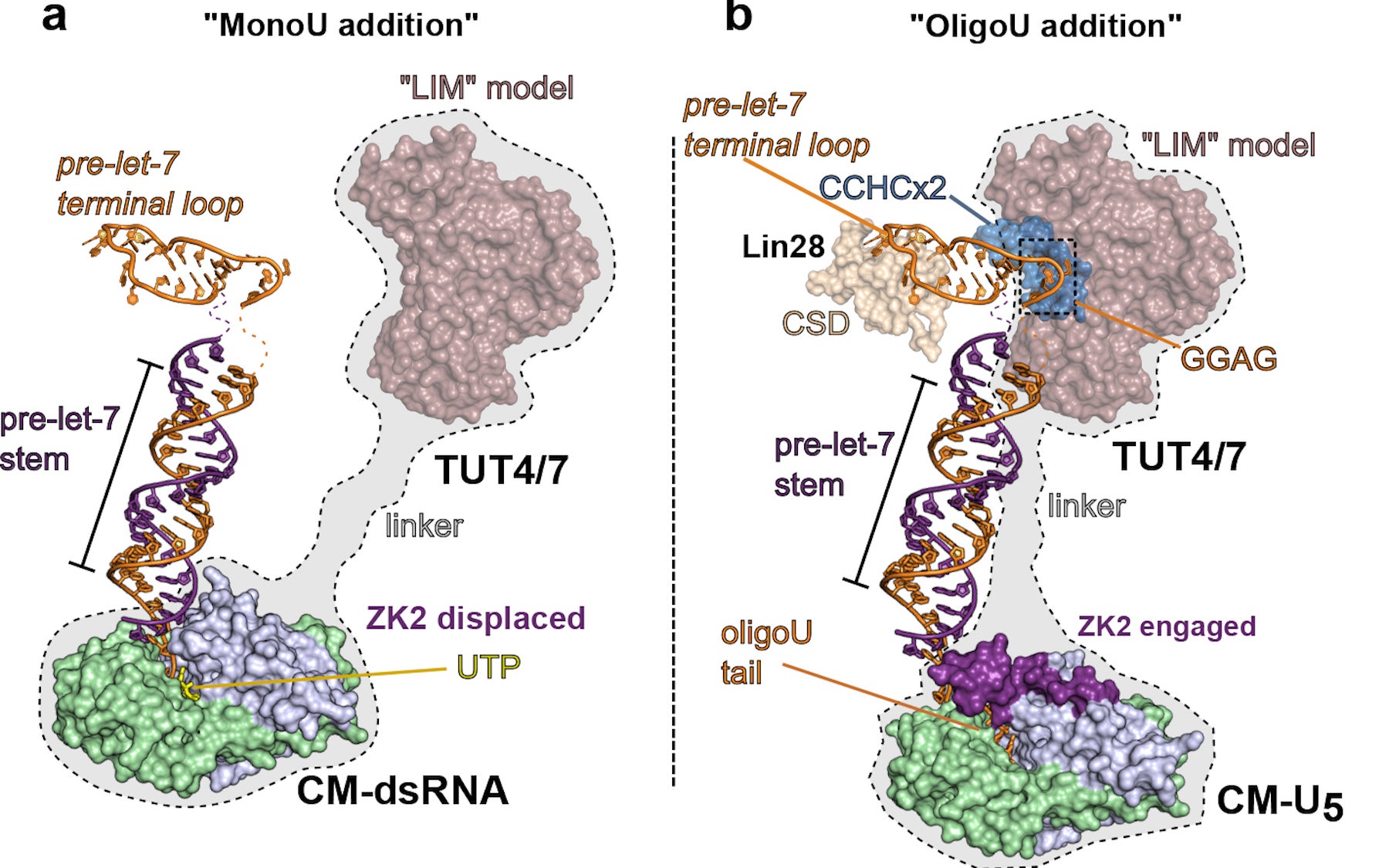
Led by CSHL Professor and Howard Hughes Medical Institute Investigator Leemor Joshua-Tor, the team used x-ray crystallography to capture the equivalent of freeze-frames of a class of enzymes called TUTases. The pictures, at the resolution of individual atoms, shows them interacting with other molecules to regulate the activity of an important small RNA molecule called let-7.
Let-7, a microRNA, is a regulator of genes important in development. When expressed in humans, it’s part of the process in which our stem cells differentiate into the various specialized cell types that populate our organs. In contexts in which it’s important for stem cells to maintain their “stem-ness”—their ability to give rise indefinitely to more stem cells—let-7 needs to be deactivated.
Let-7 is of special interest because its deactivation in cancer cells is thought by some to confer upon those cells the stem cell-like property of being able to proliferate indefinitely. This has focused attention on understanding the mechanisms regulating whether let-7 is active or inactive.
The team’s pictures show how TUT4 and TUT7—two of the TUTase enzymes—act differently in the two contexts. In a differentiating cell, the TUTases correct a small genetically induced defect in the sequence of many let-7 precursors, enabling mature let-7 microRNAs to be generated. Those defects are ameliorated when a TUTase adds a single uridine (“U”) molecule to the sequence of a let-7 precursor.
In a stem cell destined to retain its stem-ness, one of the TUTases adds not one but 30 uridines to the let-7 precursor, in this way marking it for destruction by another enzyme. “We have been able to capture the TUTases in these two different modes of action,” says Joshua-Tor.
Joshua-Tor explains that TUTases are promising targets for new cancer therapies. In kidney and lung cancer cells, for example, in which let-7 levels are far below normal, TUTases involved in marking let-7 for destruction could perhaps be inhibited. Having extremely high-resolution images of how they act in the molecular dance should help in that effort.
The team included first author Christopher Faehnl, a CSHL Research Investigator, and former Watson School of Biological Sciences student Jack Walleshauser.
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
The work was supported by NIH grant R01-GM114147; the Cold Spring Harbor Laboratory Women in Science Award; and the Watson School of Biological Sciences.
Citation
“Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis” appears July 3, 2017 in Nature Structural and Molecular Biology. The authors are: Christopher R. Faehnle, Jack Walleshauser and Leemor Joshua-Tor. The paper can be accessed at: http://www.nature.com/nsmb/index.html
Principal Investigator

Leemor Joshua-Tor
Professor, Director of Research & HHMI Investigator
W.M. Keck Professor of Structural Biology
Cancer Center Member
Ph.D., The Weizmann Institute of Science, 1991