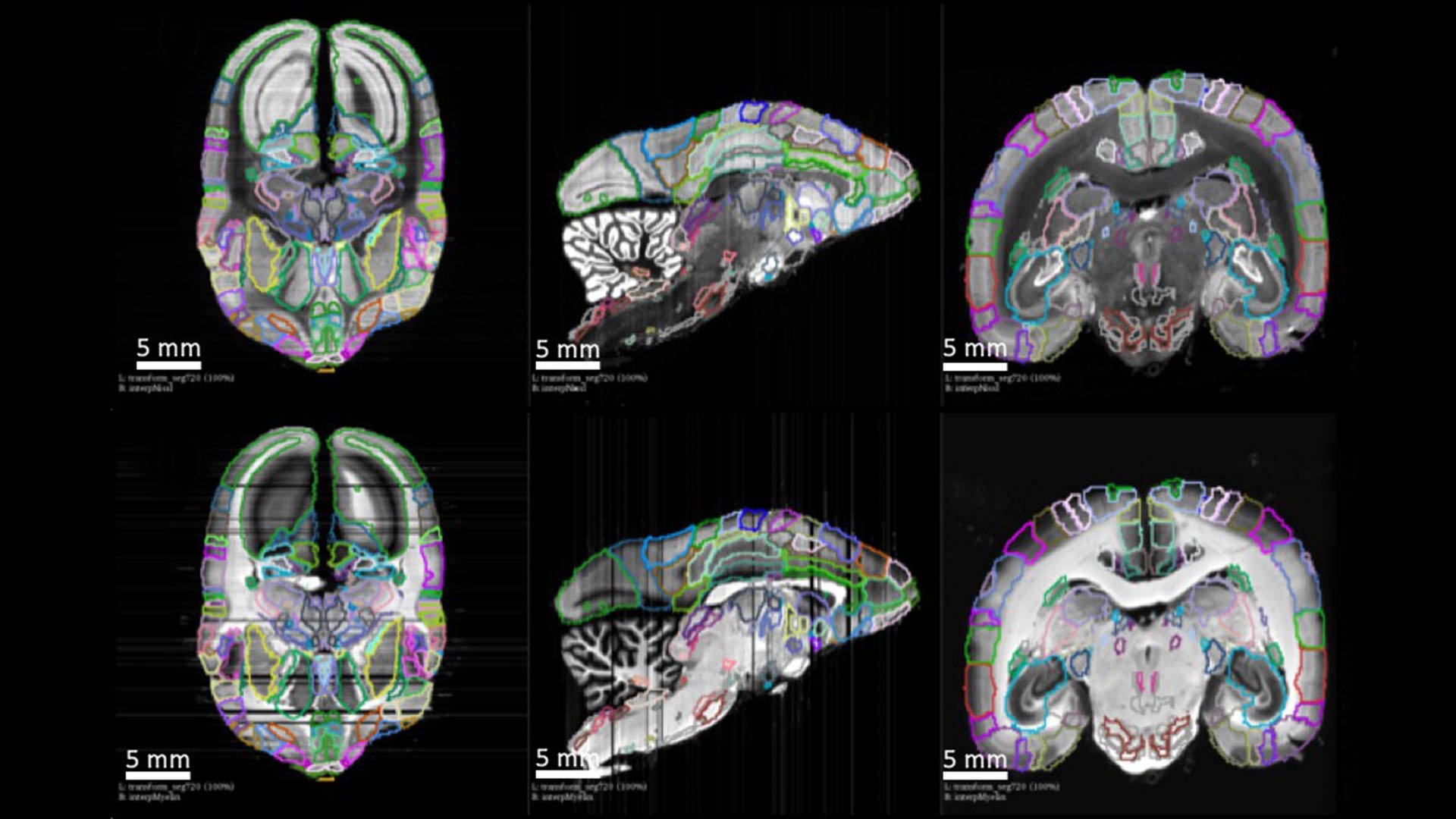
Cold Spring Harbor, NY — The ability to comprehensively map the architecture of connections between neurons in primate brains has long proven elusive for scientists. But a new study, conducted in Japan with contributing neuroscientists from Cold Spring Harbor Laboratory (CSHL), has resulted in a 3D reconstruction of a marmoset brain, as well as information about neuronal connectivity across the entire brain, that offers an unprecedented level of detail.
The study has introduced new methodology, combining experimental and computational approaches, that helps account for significant variation between individual brains. It allows for synthesizing unique brain connectivity maps into a single reference brain. The resulting data set for the marmoset brain is an ideal launching-off point for further studies, and scientists believe it may offer insights into human neural connectivity.
CSHL Professor Partha Mitra, who conceptualized and collaboratively led the study as part of Brain/MINDS research conducted at the RIKEN Center for Brain Science in Japan, explains that the endgame for any large-scale brain study is to learn more about human brain architecture and how disease can affect it. To do so, scientists must study a brain that is similar to a human’s.
The brain architecture of marmosets more closely resembles that of humans than does the mouse brain, which has been the focus of similar efforts in the past. While mice are currently the mainstay for modeling human disease, the emergence of marmoset models of human neurological disorders has made marmosets a target of new research.
Among primates, the marmosets’ relatively small brains lend themselves to thorough mapping of neural connections. And in comparison with extensively studied primates like the macaque, marmosets can be easier to study because their brain surfaces are flatter than the more folded cortical surfaces of larger primates.
The results of Mitra and colleagues’ new study are detailed in the journal eLife.
A brain fully reconstructed using MRI guided registration with process and cell detection. A clear pathway is marked by a green tracer traveling from region to region in this 3d visualization of neural projections in the marmoset brain. Courtesy of eLIFE https://doi.org/10.7554/eLife.40042.011
“Brain connectivity studies have been carried out in the marmoset before,” Mitra explains. “But we did not have complete three-dimensional digital data sets, showing connectivity patterns across several entire brains at the light-microscope resolution. The data we now have is completely unprecedented in scale and in information content.”
With this new data and approach as a basis, Mitra and other neuroscientists are one step closer to making sense of the complex neural connections in the primate—and human—brain. The hope is that this line of research will eventually lead to fundamental therapeutic advances for human diseases.
Written by: Brian Stallard, Content Developer/Communicator and Sarah Kain, Editorial Content Manager, Public Affairs | publicaffairs@cshl.edu | 516-367-8455
Funding
This research was funded by Brain Mapping of Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from the Japan Agency for Medical Research and Development, AMED, the Crick-Clay Professorship (CSHL), the Mathers Foundation, the H.N. Mahabala Chair at IIT Madras, the ARC Centre of Excellence for Integrative Brain Function, and Monash University.
Citation
Lin, M.K. et al., “A high-throughput neurohistological pipeline for brain-wide mesoscale connectivity mapping of the common marmoset” was published in eLife Sciences on February 21, 2019.