Atomic structure of human Argonaute-2 protein bound to a microRNA ‘guide’ could lead to better understanding of RNA interference mechanisms
Cold Spring Harbor, NY — In a study published in the journal Cell on May 24, Cold Spring Harbor Laboratory (CSHL) scientists describe the three-dimensional atomic structure of a human protein bound to a piece of RNA that “guides” the protein’s ability to silence genes. The protein, Argonaute-2, is a key player in RNA interference (RNAi), a powerful cellular phenomenon that has important roles in diverse biological processes, including an organism’s development.
Detailed knowledge of the structure of human Argonaute-2 and the way it interacts with its RNA guides will greatly improve our understanding of its biological mechanism of action,” says CSHL Professor and HHMI Investigator Leemor Joshua-Tor, Ph.D., the study’s leader. “Such precise structural information of the human Argonaute bound to an important RNA guide could potentially aid both basic research to understand the function of genes and also advance the development of RNAi as a therapeutic strategy in clinical settings.”
Upon the activation of a gene within a cell, the gene’s DNA is copied into a messenger RNA (mRNA) “transcript.” The instructions encoded within this transcript are then used as a blueprint by the cell’s protein synthesis machinery to generate a working protein. The gene is “silenced” or prevented from giving rise to the protein, however, when an Argonaute-2 protein that is bound to a small piece of “guide” RNA—either a short-interfering RNA or a microRNA—intercepts the mRNA molecule. The guide RNA, whose nucleotide sequence matches that of the target mRNA, acts as a homing device that helps the Argonaute-2 protein zero in on the mRNA target.
A few years ago, Joshua-Tor collaborated with CSHL Professor and HHMI Investigator Gregory Hannon, Ph.D., who is also a co-author in this study, to show that Argonaute proteins, which are made up of different domains or parts, act like a pair of molecular scissors that slice up target mRNAs, thus preventing proteins from being made and enforcing the silencing of their genes. The discovery of the Argonautes’ “slicer” activity stemmed in part from solving the crystal structure of an Argonaute protein from Pyrococcus furiosus, an archebacterium that thrives in extremely high temperatures.
“But we still know nothing about the biological functions or mechanisms of action of archebacterial Argonautes,” says Joshua-Tor. “We therefore next focused on solving the structures of Argonautes from higher organisms such as mammals, in which Argonaute functions and target recognition are well documented.”
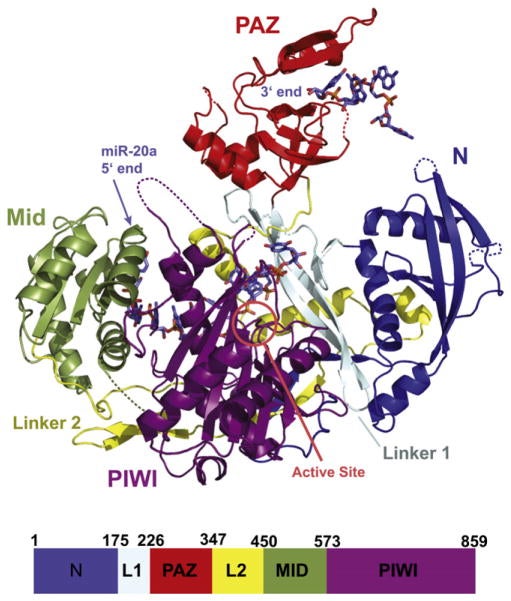
Joshua-Tor’s team and other research groups subsequently determined the atomic structures of individual parts of Argonaute proteins from higher organisms. While these studies revealed several important details—for example, the interaction between two parts of the Argonaute protein, called the PAZ and Mid domains, with the two ends of guide RNAs—Joshua-Tor’s goal was to solve the structure of the entire human Argonaute protein in complex with a single human guide RNA.
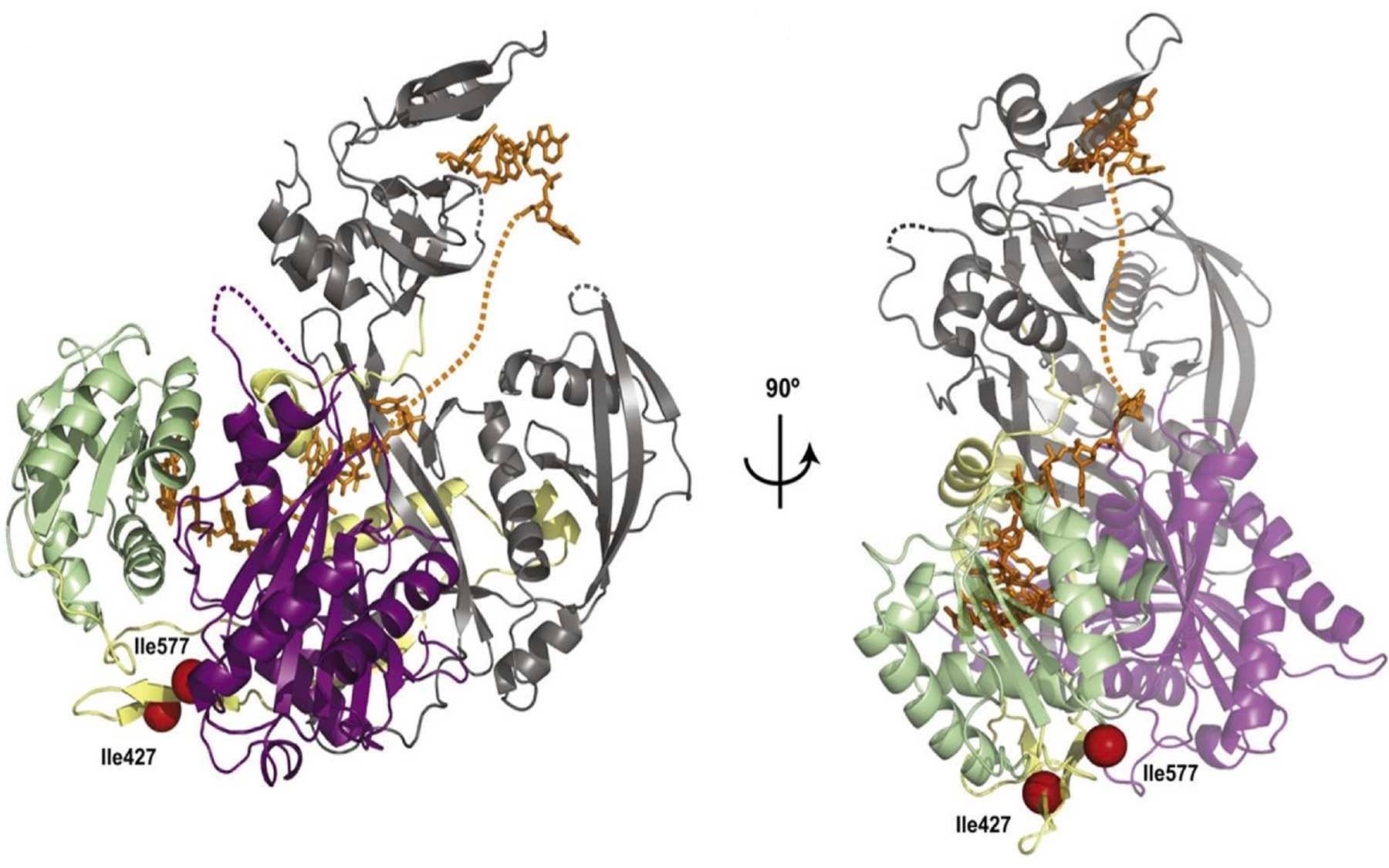
Overcoming a complicated series of technical challenges, her team has achieved this goal by analyzing the structure of a full-length human Argonaute-2 protein bound to a small RNA called miR-20a, which is known to play a role in cancer development. Although Argonautes from higher organisms diverged from their archebacterial cousins more than three billion years ago, the team’s analysis shows a remarkable similarity between the two structures, especially in the regions that are important for target recognition and slicing activity.
“Our structure shows that the guide RNA, which is anchored at both ends by the PAZ and Mid domains, kinks and twists its way through the structure of the entire protein, making several points of contact within each domain and with the linker loops that join them,” explains Joshua-Tor. “The guide RNA thus acts like a backbone that rigidly locks together the otherwise flexible Argonaute protein and gives it stability.”
The researchers speculate that the path threaded through the Argonaute by the guide RNAs could have evolved to maximize mutual stability, in turn making the protein-RNA complexes long-lived. This long life is critical for many biological processes that are mediated by Argonautes. “This is also the kind of information that might help us to design better synthetic guide RNAs for therapeutic use,” explains Joshua-Tor. “It will also be useful to researchers who are trying to find more precise ways of blocking Argonaute activity.”
Written by: Hema Bashyam, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
The work was supported by the Louis Morin Charitable Trust and by a grant (GM062534) from the National Institutes of Health. Leemor Joshua-Tor and Greg Hannon are Howard Hughes Medical Institute Investigators.
Citation
“The structure of human Argonaute-2 in complex with miR-20a” appeared online in Cell on May 24. The full citation is: Elad Elkayam, Claus-D. Kuhn, Ante Tocilj, Astrid D. Haase, Emily M. Greene, Gregory J. Hannon, and Leemor Joshua-Tor. The paper can be downloaded at www.cell.com using the doi 10.1016/j.cell.2012.05.017.