Cold Spring Harbor, NY — RNA interference (RNAi) has emerged as a fundamentally important biological phenomenon and as a versatile, powerful tool for biomedical research. In organisms from fungi and flies to plants and mammals, RNAi plays a multifaceted role in molecular biology by silencing genes through chromatin remodeling, interfering with protein synthesis, and—in its best-studied mode of action—quashing gene expression by cleaving messenger RNA. Experimental applications of RNAi have spurred the exploration of gene function in many basic research, drug discovery, and clinical settings. Until now, however, the identity of the molecular scissors that carry out RNAi-mediated messenger RNA cleavage has not been revealed.
Two studies published this week in Science have resolved this mystery by establishing that Argonaute2, a signature protein component of the RNA interference machinery, provides the cutting action that carries out RNAi-mediated messenger RNA cleavage. The studies were conducted at Cold Spring Harbor Laboratory by research groups led by Greg Hannon and Leemor Joshua-Tor.
Given the role of RNA interference in a wide variety of biological processes and the intense interest in RNAi as a tool for both basic and applied research, the studies by Hannon and Joshua-Tor are a significant advance toward a comprehensive understanding of one of the most intriguing and far-reaching biological phenomena to be uncovered in recent years.
Full Story
Hannon’s group focused on sorting out the functions of four mammalian Argonaute family members (Ago1, 2, 3, and 4). First, through a biochemical approach, Hannon and his colleagues found that only a single Argonaute family member, Ago2, supports the formation of mRNA cleavage-competent complexes in vitro.
To extend these biochemical findings to an in vivo setting, and to further explore the specialization of Argonaute family member function, Hannon’s group disrupted the mouse Ago2 gene by targeted insertional mutagenesis. The researchers observed an embryonic lethal phenotype and striking developmental abnormalities in Ago2 homozygotes.
All Ago2 homozygous embryos displayed defects in neural tube structure, with half of the embryos showing complete failure of neural tube closure in the head region. The embryos also had enlarged hearts and pronounced swelling of the pericardial cavity, and were severely developmentally delayed compared to their wild-type and heterozygous littermates. In contrast to the critical developmental role of Ago2 revealed by these findings, work by researchers elsewhere has shown that other Argonaute family members are dispensable for development.
Hannon and his colleagues reasoned that if Ago2 is uniquely capable of assembling into mRNA cleavage-competent complexes in vivo, then mouse cells lacking Ago2 but containing other Ago proteins should be incapable of carrying out experimentally-triggered, siRNA-mediated mRNA cleavage.
The researchers observed just such a result with Ago2-deficient mouse cells, and showed further that a plasmid encoding human Ago2 (but not Ago1) could restore siRNA-mediated mRNA cleavage to Ago2-deficient mouse cells.
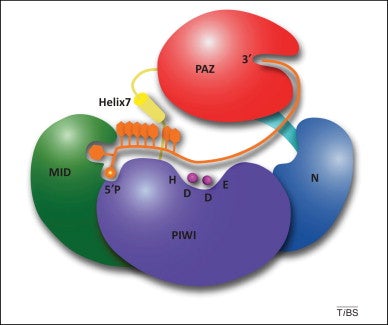
Hannon’s study was consistent with the possibility that Argonaute2 provides the “Slicer” activity of RNAi-mediated mRNA cleavage. However, the possibility that a different protein provides Slicer activity could not be ruled out. Fortunately, Leemor Joshua-Tor’s group, also at Cold Spring Harbor Laboratory, was simultaneously conducting x-ray crystallographic studies of an Argonaute protein (PfAgo) from the hyperthermophilic archaebacterium, Pyrococcus furiosus.
When Joshua-Tor and her colleagues determined the crystal structure of PfAgo, they found that the PIWI domain of PfAgo belongs to the RNase H family of enzymes, whose members have RNA cleavage activity. This finding immediately implicated Argonaute itself as the protein that provides the Slicer activity of RNAi in mammals and other organisms.
Hannon and Joshua-Tor had previously shown that another domain of Argonaute proteins, the PAZ domain, recognizes the 3′ ends of siRNAs. That observation, combined with her group’s discovery that the PIWI domain of PfAgo belongs to the RNase H family, enabled Joshua-Tor to propose a model for siRNA-targeted mRNA cleavage by Argonaute.
Collectively, the two new studies establish that Argonaute2 is the catalytic engine of mammalian RNAi and provide a specific, structural mechanism for siRNA-targeted mRNA cleavage by Argonaute.
*The term “Argonaute” refers to the squid-like appearance of the leaves of Arabidopsis mutants lacking AGO1 gene function (Bohmert et al., 1998). See also the Greek myth Jason and the Argonauts and the squid, Argonauta argo.
Written by: Communications Department | publicaffairs@cshl.edu | 516-367-8455