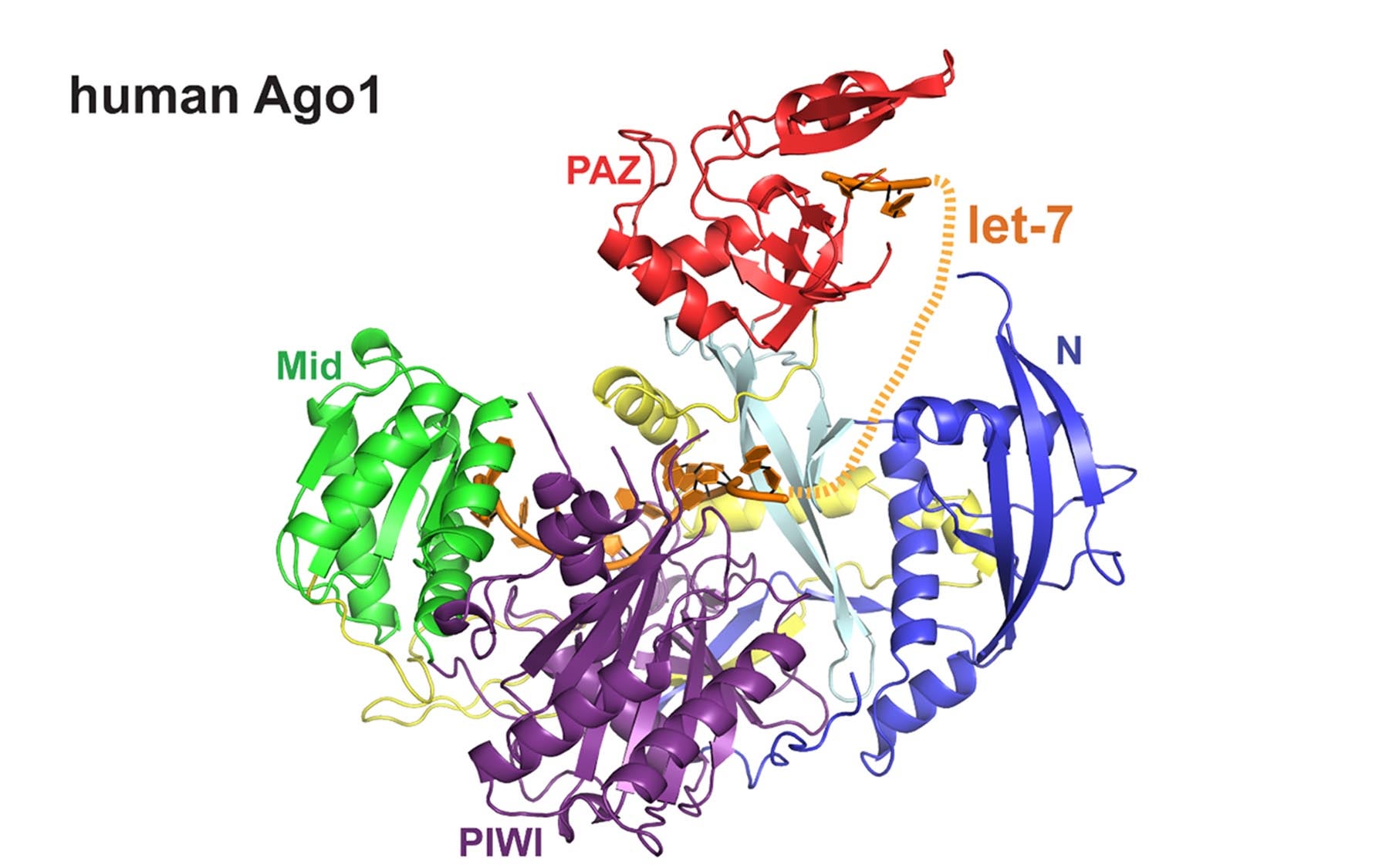
Human Argonautes (hAgo), are key proteins involved in a process known as RNA interference
Cold Spring Harbor, NY — Human Argonautes (hAgo), are key proteins involved in a process known as RNA interference. RNAi, as it is often called, is a mechanism that cells use to regulate gene expression. Human Argonaute-2 (hAgo2) is known as “slicer” for its unique ability among the 4 human Argonaute proteins to directly cut messenger RNA—which carries the information coded in genomic DNA to make a protein—and thus disable “messages” generated from genes.
The atomic resolution structure of hAgo2 solved previously, revealed the active site, a region of the enzyme that is key to its slicing activity. The other human Argonaute proteins, hAgo1, 3 and 4, while very similar to hAgo2, lack slicing activity. So an important question left unanswered was: what features in these proteins explain why none of them are able to act as “slicers”?
A team at Cold Spring Harbor Laboratory (CSHL) led by Professor and HHMI Investigator Leemor Joshua-Tor of the W. M. Keck structural biology laboratory today publishes a paper in the journal Cell Reports that defines the critical differences between the human Argonautes that lead to their differences in activity.
Here, the group determined the structure of the non-slicer hAgo1 in complex with a microRNA (miRNA) called let-7. This RNA does not code for a protein but is highly conserved by evolution, occurring in many species, and is involved in regulating gene expression during human development and cancer. “This is the highest-resolution structure of a eukaryotic Argonaute to date, which allows for more in-depth analysis of important features like its active site,” says Joshua-Tor.
However, the non-slicer Argonaute looks very similar to its catalytically active counterpart. The group therefore used mutational analysis to pinpoint the source of the differences in activity among the human Argonaute proteins. The team demonstrates in their new work the ability to transform hAgo1 into a “slicer” by changing key parts of the protein at and away from the active site.
Identification of previously unrecognized elements involved in ‘slicer’ activity
At the heart of the active site of hAgo2 are 4 amino acids, the building blocks of proteins. In hAgo1, Joshua-Tor and her team, which also included CSHL Professor and HHMI Investigator Greg Hannon, found a single amino acid change in this key sequence. But when the sequence was experimentally altered to match that of the “slicer” Argonaute hAgo2, hAgo1 still proved unable to function as a slicer. This clearly indicated that other factors in the structure of hAgo2 are essential to its RNA-slicing activity.
“We looked for other mutations and found that an additional mutation was needed to activate hAgo1 slicing,” says Chris Faehnle, the first author of the paper. This additional mutation is located in an area of the structure where it binds to RNA, known as the RNA binding groove. Although this mutation, along with the other amino acid change, enabled hAgo1 to act as a slicer, it was still not as efficient as hAgo2.
In a final experimental step, however, when Joshua-Tor’s team swapped in the so-called N-domain region of hAgo2 for the corresponding region in hAgo1, slicing activity was enhanced. The same result was also obtained for the slicer-inactive hAgo3. This means that despite being extremely similar in structure, there are mutations and entire regions of the hAgo2 protein that are optimized for slicing activity, which are not present in hAgo1, 3, and 4.
The implication: It is not just the active site that is important for determining the activity of a protein. Other protein regions can contribute to the activity of the whole through subtle changes in the overall structure.
“This helps us understand exactly how these proteins work at the molecular level, which is important for the design of new and improved therapeutics that use small RNAs,” said Joshua-Tor.
Written by: Edward Brydon, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
The research described in this release was supported by the following grants and funding agencies: The Louis Morin Charitable Trust and the Robertson Research Fund of Cold Spring Harbor Laboratory (to L. J.). G. J. H. and L. J. are investigators of the Howard Hughes Medical Institute.
Citation
“The Making of a Slicer:Activation of Human Argonaute 1” is published online in Cell Reports on June 6, 2013. The authors are: Christopher R. Faehnle, Elad Elkayam, Astrid D. Haase, Gregory J. Hannon, and Leemor Joshua-Tor. The paper can be obtained online at doi:10.1016/j.celrep.2013.05.033.
Principal Investigator

Leemor Joshua-Tor
Professor, Director of Research & HHMI Investigator
W.M. Keck Professor of Structural Biology
Cancer Center Member
Ph.D., The Weizmann Institute of Science, 1991