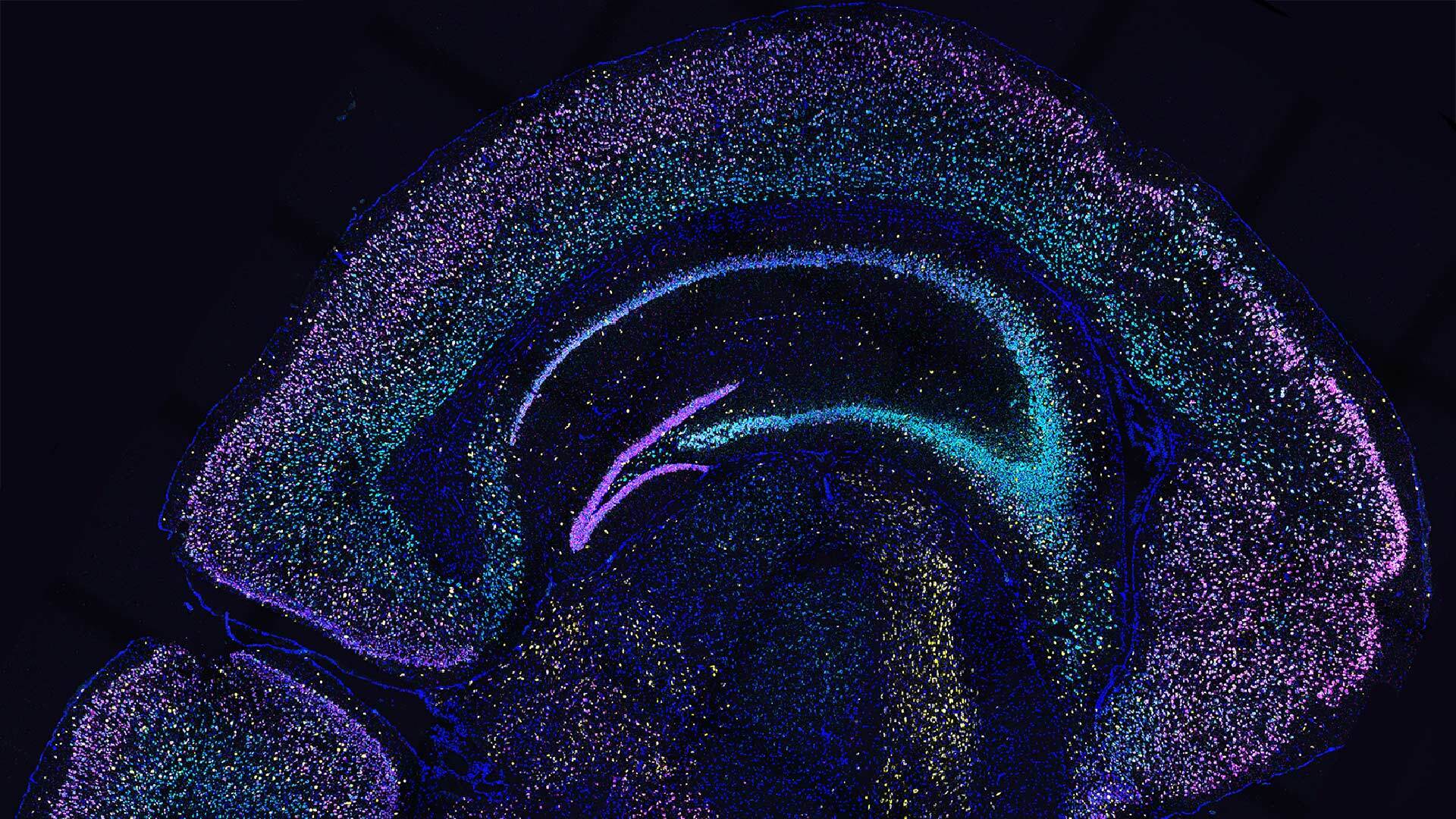
CSHL Professor Anthony Zador has developed mapping technologies that identify connections of individual brain cells, pinpoint where a cell is located, and determine the specific function of that cell. Detailed wiring diagrams—connectomes—for the brain are critical for understanding brain development, function, and disease.
These technologies, called MAPseq and BARseq, use genetic barcodes, or short sequences of DNA and RNA, to trace thousands of brain circuits simultaneously. The approach increases the number of neuronal connections that can be traced at a time versus traditional microscopy-based methods that rely on fluorescent markers or dyes to trace neuronal activity. Zador’s genome sequencing technology approach allows researchers to label many neurons and brain regions at once.
MAPseq labels each neuron with a unique barcode so that researchers can determine where a neuron projects by dissecting brain regions of interest and searching for the barcode. BARseq is the next generation of MAPseq, allowing barcodes to be sequenced within a tissue, providing a method for in situ sequencing in neurons. This approach preserves anatomical information and allows scientists to examine cellular connectivity at greater resolution, which has several possible applications including cell typing and brain mapping.
Written by: Dagnia Zeidlickis, Vice President, Communications | zeidlick@cshl.edu | 516-367-8455