Cold Spring Harbor, NY — Because liver cancer is particularly diverse, genetically, and prone to relapse, identifying biomarkers that can predict disease progression is a critical goal in the fight against it.
Researchers at Cold Spring Harbor Laboratory (CSHL), led by CSHL Professor Adrian Krainer, now report in Genome Research that they have developed a method for identifying splicing-based biomarkers for the most common form of liver cancer, hepatocellular carcinoma (HCC). They believe the method will be useful in other cancer types as well.
“This study underscores the potential for learning how RNA splicing variants can contribute to cancer and points to these variants as potential biomarkers for cancer progression,” Krainer says.
Splicing refers to a process in which an RNA message copied from information encoded in a gene is edited before it is able to serve as a blueprint for the manufacture of a specific protein. A gene can give rise to multiple RNA messages, each resulting in a different protein variant, or “isoform.” Many diseases have been associated with errors or variations in the way that RNA is spliced. Errors or variations in splicing can lead to non-functional proteins or proteins with distinct or aberrant functions.
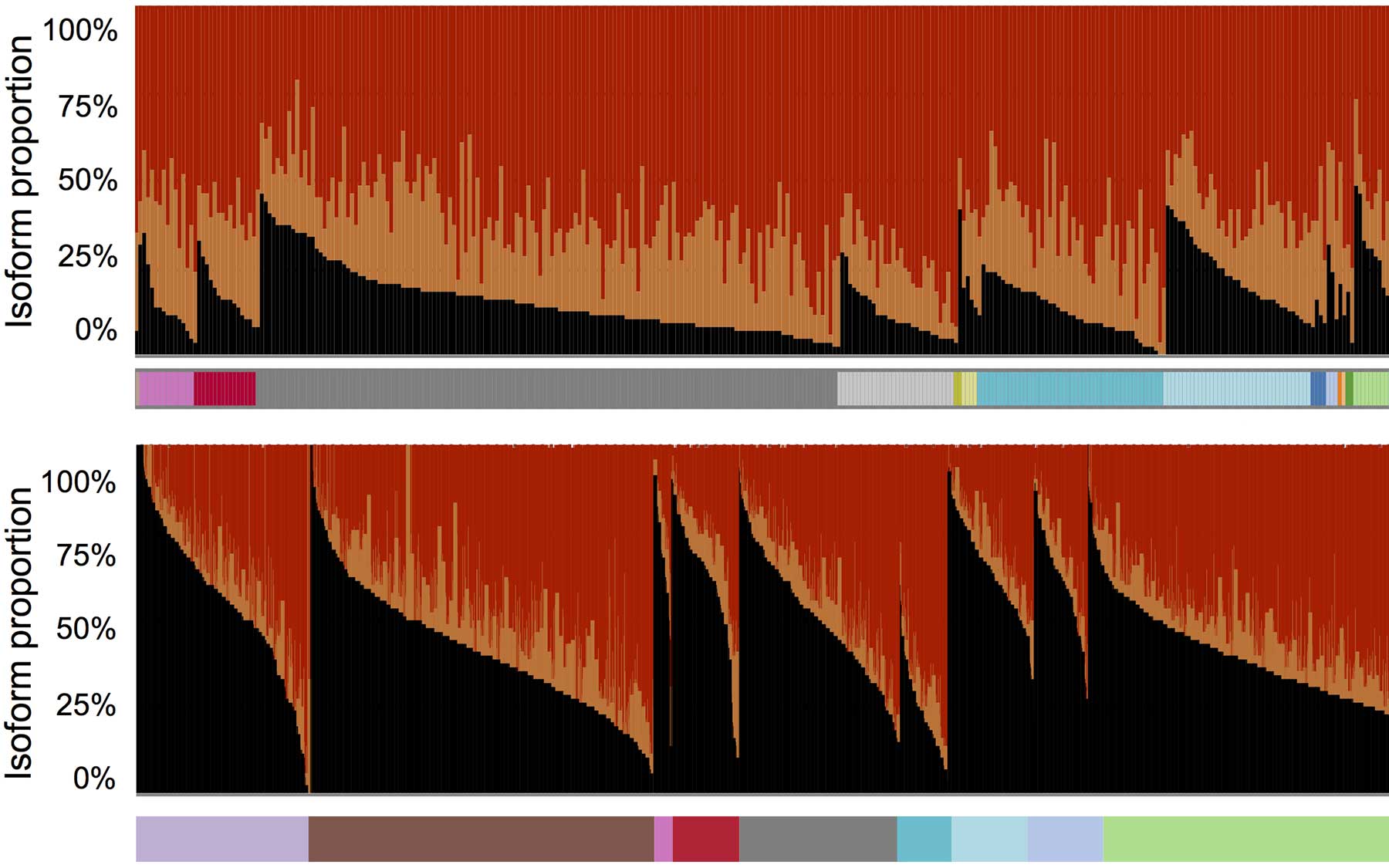
Recent studies have identified splicing irregularities in liver cancer cells. Led by CSHL postdoctoral researcher Kuan-Ting Lin, Krainer’s team developed a method that comprehensively analyzes all RNA messages made from a given gene. The team tested their splicing-variant detection method in HCC, by analyzing RNA messages in HCC cells sampled from hundreds of patients.
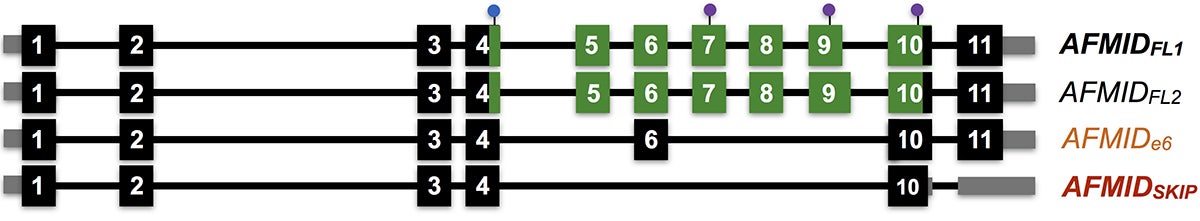
They found that particular splicing isoforms of the gene AFMID correlated with very poor patient survival. These variants lead cells to manufacture truncated versions of the AFMID protein. These unusual versions of the protein are associated in adult liver cancer cells with mutations in tumor-suppressor genes called TP53 and ARID1A.
These mutations, the researchers hypothesize, are associated with low levels of a molecule called NAD+ that is involved in repairing damaged DNA. Restoring missing portions, called exons, to AFMID’s normal RNA message, they propose, might raise NAD+ to normal levels, avoiding mutations in TP53 and ARID1A. The team hopes to use small molecules called ASOs (antisense oligonucleotides) that can bind to RNA, to change the way AFMID’s RNA messages are spliced. Krainer’s team previously used this technique to correct errors in the splicing of the gene SMN2 as a way to treat spinal muscular atrophy (SMA).
Fixing AFMID splicing could lead to enhanced production of NAD+ and an increase in DNA repair. “If we can do this, AFMID splicing can become a therapeutic target and the source of a new drug for liver cancer,” Lin says. Preliminary experiments suggest the team is on the right track. They demonstrated that coaxing cells to over-express AFMID spliced in the normal manner led to higher NAD+ levels and slower growth of liver cancer cells.
Written by: Chris Palmer, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
National Institutes of Health
Citation
Lin, K-T et al, “A human-specific switch of alternatively spliced AFMID isoforms contributes to TP53 mutations and tumor recurrence in hepatocellular carcinoma,” appeared online in Genome Research February 15, 2018.
Principal Investigator

Adrian R. Krainer
Professor
St. Giles Foundation Professor
Cancer Center Program Co-Leader
Ph.D., Harvard University, 1986