A single gene is identified whose regulation controls oil palm yield
Kuala Lumpur, Malaysia, St. Louis, MO and Cold Spring Harbor, NY — A multinational team of scientists has identified a single gene, called Shell, that regulates yield of the oil palm tree. The fruit and seeds of the oil palm are the source of nearly one-half of the supply of edible vegetable oil worldwide, and provide one of the most promising sources of biofuel.
The discovery, the product of a multiyear effort to provide a high-quality full genome map of the oil palm plant and to scour the sequence for genes of importance to both science and industry, has major implications for agriculture and the environment.
“The discovery that regulation of the Shell gene will enable breeders to boost palm oil yields by nearly one-third is excellent news for the rainforest and its champions worldwide,” says Datuk Dr. Choo Yuen May, the Director General of the Malaysian Palm Oil Board (MPOB), an agency of the Malaysian federal government.
The discovery was made by researchers at the MPOB in conjunction with scientists at St. Louis-based Orion Genomics. Also lending support were scientists in New York, at Cold Spring Harbor Laboratory (CSHL) and the American Museum of Natural History. The international team’s work is detailed in two papers published online today in Nature.

“The discovery of Shell indicates a clear path toward more intensive use of already planted lands, and thus should lessen pressures to expand the land area devoted to oil palm, notably onto endangered rainforest land—a major concern for the environment and a rallying point for activists in recent years,” says Robert A. Martienssen, Ph.D., scientific co-founder of Orion Genomics, who is also a professor of plant genetics at CSHL.
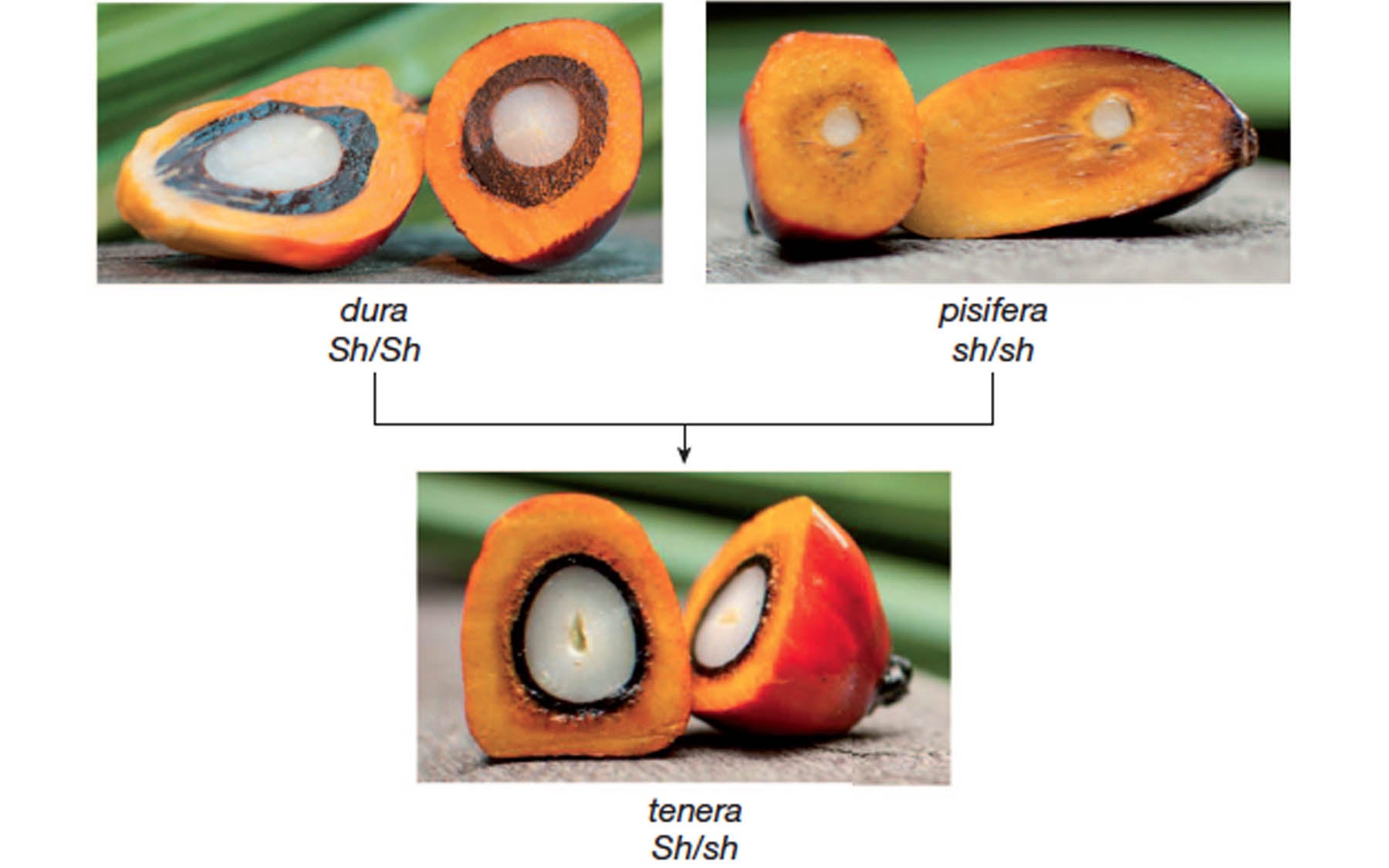
“Mutations in Shell explain the single most important economic trait of the oil palm: how the thickness of its shell correlates to fruit size and oil yield,” explains Dr. Rajinder Singh of the MPOB, first author of the Nature paper describing the Shell gene.
There are two species of oil palm, African (Elaeis guineensis) and South American (Elaeis oleifera). Together they account for 45 percent of the edible vegetable oil worldwide. Palm oil also has the best energy balance of any commercial product currently used in biofuel applications, yielding about 9 times the energy required to produce it, according to Dr. Martienssen.
The Shell gene is responsible for the oil palm’s three known shell forms: dura (thick); pisifera (shell-less); and tenera (thin), a hybrid of dura and pisifera palms. Tenera palms contain one mutant and one normal version, or allele, of Shell, an optimum combination that results in 30% more oil per land area than dura palms.
How the discovery will affect plantation management and land use
The discovery of the Shell gene and its two naturally occurring mutations highlight new molecular strategies to identify seeds or plantlets that will become high-yielding palms before they are introduced into plantations.
Seed producers can now use the genetic marker for the Shell gene to distinguish the three fruit forms in the nursery long before they are field-planted. Currently, it can take six years to identify whether an oil palm plantlet is a high-yielding palm. Even with selective breeding, 10 to 15 percent of plants are the low-yielding dura form due to uncontrollable wind and insect pollination, particularly in plantations without stringent quality control measures.
“Accurate genotyping for enhanced oil yields will optimize and help stabilize the acreage devoted to oil palm plantations, providing an opportunity for the conservation of rainforest reserves,” Martienssen explains.
The Malaysian government strongly supported the genome sequencing project for the nation’s most important crop. The government halted the conversion of new forest land for agriculture, including palm oil, in the 1990s. According to the MPOB, the government has committed to preserve 50 percent of Malaysia’s total land area as forest. To meet increased demand for palm oil, the government converted colonial rubber and cocoa plantations to oil palm plantations.
What the full genome sequences reveal

The discovery of Shell occurs in the context of a broader effort to map the genomes of both the African and South American oil palm species. One of the newly published maps is the 1.8 gigabase sequence of the E. guineensis African oil palm. It comprises nearly 35,000 genes, including the full set of oil biosynthesis genes and other transcriptional regulators highly expressed in the oil-rich palm fruit.
The researchers also created a draft sequence of the South American oil palm E. oleifera. Both palm species are in the Arecaceae family of flowering plants, which fossil evidence dates to the Cretaceous period, an estimated 140 to 200 million years ago. The investigators’ comparison of the two maps enabled them to estimate that the oil palm species diverged at the old world—new world split.
For further information on the oil palm genome project, please visit http://genomsawit.mpob.gov.my
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Citation
“Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds” and “The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK” will be published online ahead of print in Nature on Wednesday, July 24, 2013. The papers can be viewed then at: http://www.nature.com/nature/index.html
About Oil Palm
Oil palm is the most productive oil-bearing crop. Planted on only five percent of the total vegetable oil acreage, palm oil accounts for 33 percent of vegetable oil, and 45 percent of edible oil worldwide. In addition to its application as a food oil, palm oil and biomass from the oil palm has applicability as a biofuel, and biodiesel including palm oil methyl ester is in use across the globe. Like other trees, the oil palm plant serves as a natural reservoir for carbon and is more effective at sequestering carbon than other major vegetable crops. Therefore, oil palms can play a more significant role than many other crops in removing the greenhouse gas carbon dioxide from the atmosphere.
About the Malaysian Palm Oil Board
The Malaysian Palm Oil Board (MPOB) is a Malaysian government agency dedicated to serving the country’s palm oil industry. MPOB envisions the development of a highly diversified, globally competitive and sustainable oil palm industry. MPOB is focused on improving production efficiency and the quality of palm oil-derived products, optimizing land use in oil palm areas and finding new uses for palm oil derivatives. For further information, visit www.mpob.gov.my
Principal Investigator

Rob Martienssen
Professor & HHMI Investigator
William J. Matheson Professor
Cancer Center Member
Ph.D., Cambridge University, 1986