Blocking production of a pyruvate kinase splice-variant shows therapeutic promise
Cold Spring Harbor, NY — Cancer cells grow fast. That’s an essential characteristic of what makes them cancer cells. They’ve crashed through all the cell-cycle checkpoints and are continuously growing and dividing, far outstripping our normal cells. To do this they need to speed up their metabolism.
CSHL Professor Adrian Krainer and his team have found a way to target the cancer cell metabolic process and in the process specifically kill cancer cells.
Nearly 90 years ago the German chemist and Nobel laureate Otto Warburg proposed that cancer’s prime cause was a change in cell metabolism—i.e., in cells’ production and consumption of energy. In particular cancer cells have a stubborn propensity to eschew using glucose as a source to generate energy. This is known as the Warburg Effect.
While metabolic changes are an important feature in the transformation of normal cells into cancer cells they are not now thought to be cancer’s primary cause. Despite this, metabolic changes remain an attractive target for cancer therapy, as Krainer and colleagues show in a paper published online today in Open Biology, the open-access journal of Great Britain’s Royal Society.
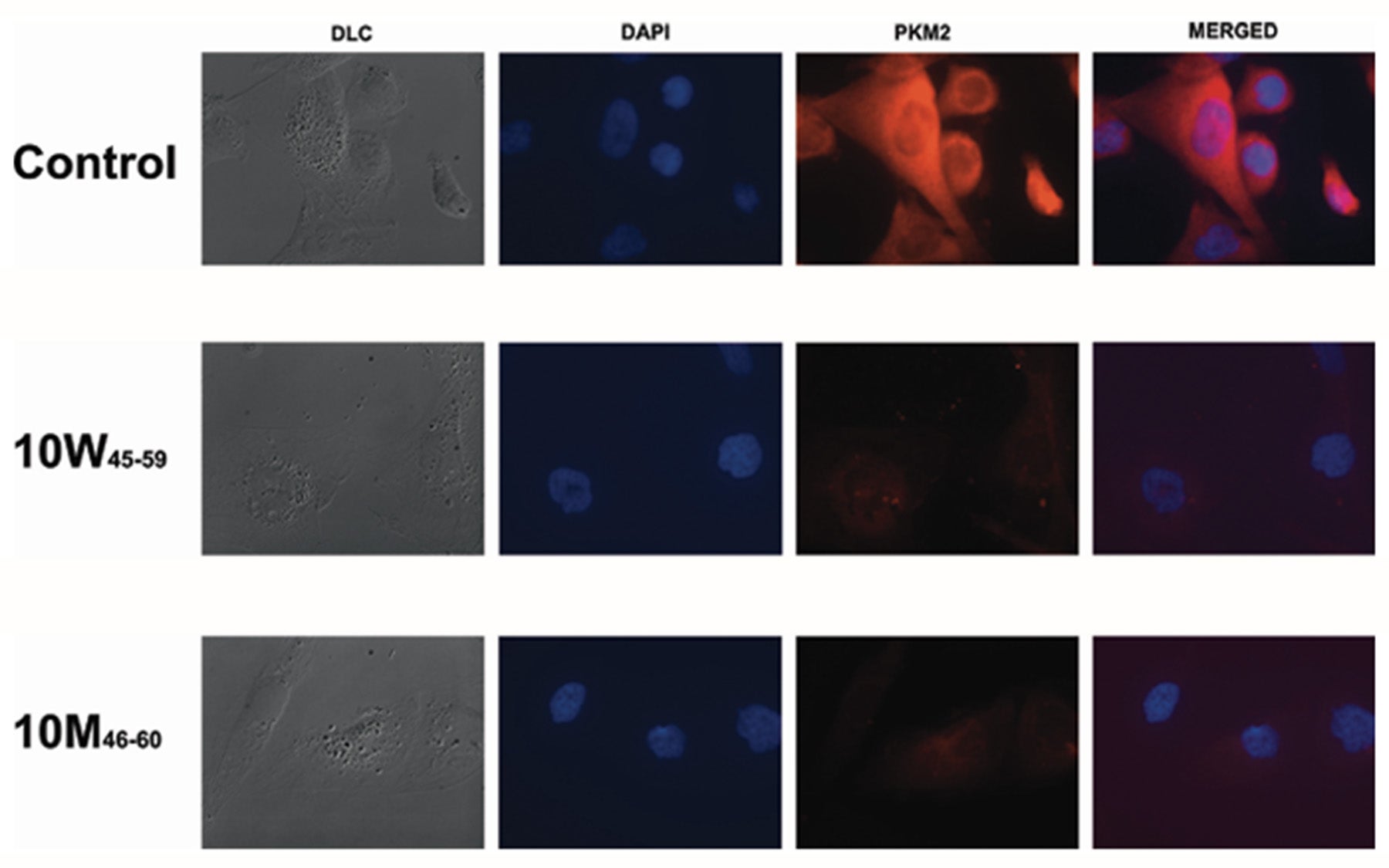
One difference between metabolism in cancer and normal cells is the switch in cancer to the production of a different version, or isoform, of a protein produced from the pyruvate kinase-M (PK-M) gene. The protein version produced in normal cells is known as PK-M1, while the one produced by cancer cells is known as PK-M2.
PK-M2 is highly expressed in a broad range of cancer cells. It enables the cancer cell to consume far more glucose than normal, while using little of it for energy. Instead, the rest is used to make more material with which to build more cancer cells.
PK-M1 and PK-M2 are produced in a mutually exclusive manner—one-at-a-time, from the same gene, by a mechanism known as alternative splicing. When a gene’s DNA is being copied into the messenger molecule known as mRNA, the intermediate template for making proteins, a cellular machine called the spliceosome cuts and pastes different pieces out of and into that mRNA molecule.
The non-essential parts that are edited out are known as introns, while the final protein-coding mRNA consists of a string of parts pasted together known as exons. The bit that fits into the PK-M1 gene-coding sequence is known as exon 9, while it is replaced in PK-M2 by exon 10. In this way alternative splicing provides the cell with the ability to make multiple proteins from a single gene.
Krainer, an authority on alternative splicing, previously published research on the protein regulators that facilitate the splicing mechanism for PK-M. His team showed that expression of PK-M2 is favored in cancer cells by these proteins, which act to repress splicing for the PK-M1 isoform. In the study published today the team explains that it decided to target the splicing of PK-M using a technology called antisense, rather than target the proteins that regulate the splicing mechanism.
Using a panel of antisense oligonucleotides (ASOs), small bits of modified DNA designed to bind to mRNA targets, they screened for new splicing regulatory elements in the PK-M gene. The idea was that one or more ASOs would bind to a region of the RNA essential for splicing in exon 10 and reveal that site by preventing splicing of exon 10 from occurring.
Indeed, this is what happened. “We found we can force cancer cells to make the normal isoform, PK-M1,” sums up Krainer. In fact, a group of potent ASOs were found that bound to a previously unknown enhancer element in exon 10, i.e., an element that predisposes for expression of the PK-M2 isoform, thus preventing its recognition by splicing-regulatory proteins. This initiated a switch that favored the PK-M1 isoform.
When they then deliberately targeted the PK-M2 isoform for repression in cells derived from a glioblastoma, a deadly brain cancer, all the cells died. They succumbed through what is known as programmed cell death or apoptosis—a process whereby the cell shuts down its own machinery and chops up its own DNA in committing a form of cellular suicide.
As to why the cells die when PK-M2 is repressed: the team found it was not due to the concomitant increase in PK-M1 (the cells survived even when extra PK-M1 was introduced). Rather, it was the loss of the PK-M2 isoform that was associated with the death of the cancer cells. How this works is still unclear but a subject of investigation in the Krainer laboratory.
The next step will be to take their ASO reagents into mouse models of cancer to see if they behave the same way there. While there are some technical and methodological obstacles to overcome, Krainer is optimistic.
“PK-M2 is preferentially expressed in cancer cells, a general feature of all types of cancer—it’s a key switch in their metabolism,” he says. Thus targeting the alternative splicing mechanism of PK-M2 using ASOs has the potential to be a cancer therapeutic with many applications.
Written by: Edward Brydon, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
The research described in this release was supported by the National Cancer Institute grant CA13106, the St. Giles Foundation, and a National Science Scholarship from the Agency for Science, Technology and Research, Singapore.
Citation
The paper can be obtained online at the following link: Zhenxun Wang, Hyun Yong Jeon, Frank Rigo, C. Frank Bennett and Adrian R. Krainer. 2012 Manipulation of PK-M mutually exclusive alternative splicing by antisense oligonucleotides. Open Biology 2: 120133. http://rsob.royalsocietypublishing.org/content/2/10/120133.full
Principal Investigator

Adrian R. Krainer
Professor
St. Giles Foundation Professor
Cancer Center Program Co-Leader
Ph.D., Harvard University, 1986