Activity of the rutabaga gene determines much of a fruit fly’s ability to learn and remember
Cold Spring Harbor, NY — Why is it that you can instantly recall your own phone number but have to struggle with your mental Rolodex to remember a new number you heard a few moments ago? The two tasks “feel” different because they involve two different types of memory—long-term and short-term, respectively—that are stored very differently in the brain. The same appears to be true across the animal kingdom, even in insects such as the fruit fly.
Assistant Professor Josh Dubnau, Ph.D., of Cold Spring Harbor Laboratory (CSHL) and his team have uncovered an important molecular and cellular basis of this difference using the fruit fly as a model. The results of their study appear in the August 25 issue of Current Biology.
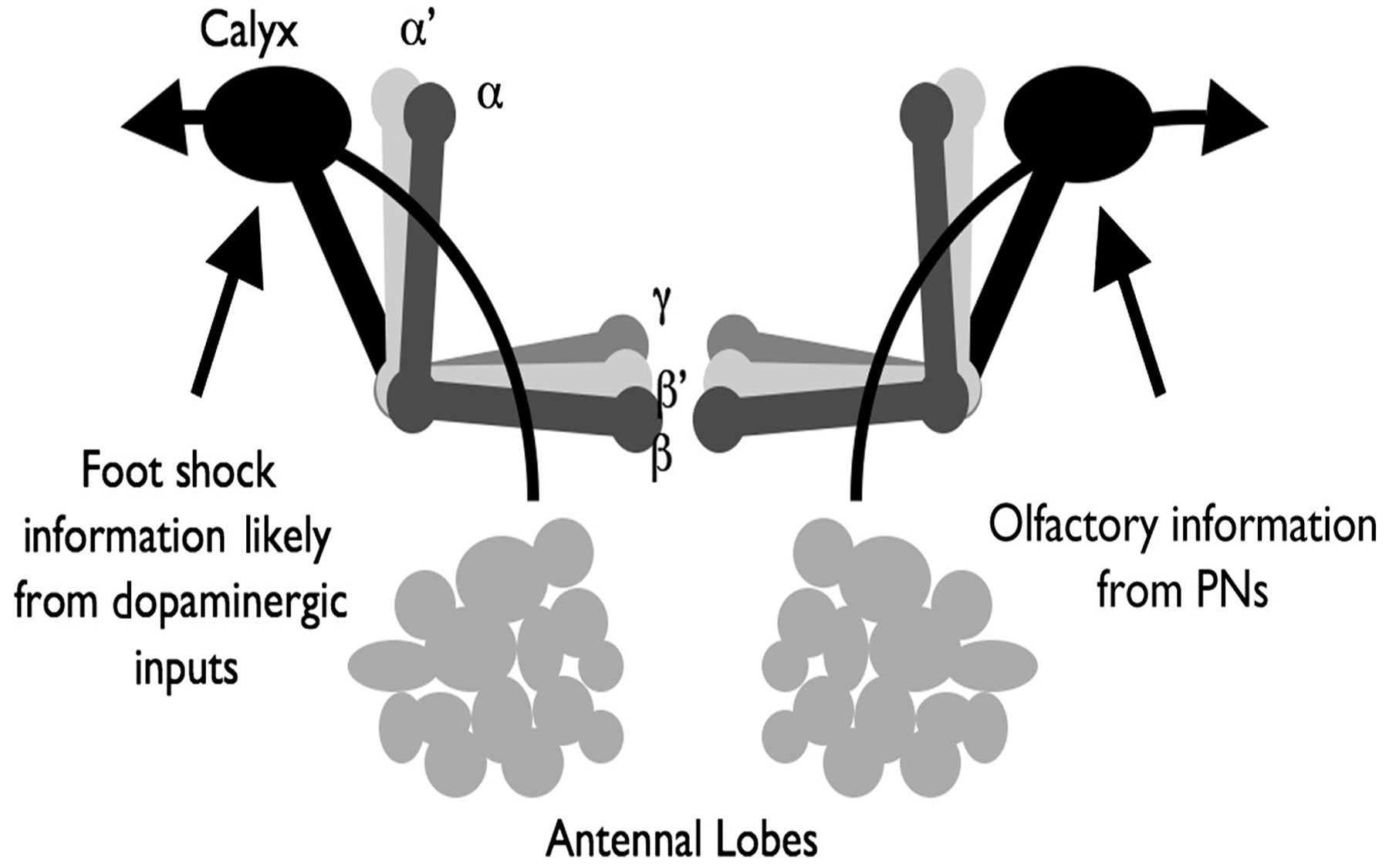
The CSHL team has found that when fruit flies learn a task, each of two different groups of neurons that are part of the center of learning and memory in the fly brain simultaneously forms its own unique memory signal or trace. Both types of trace, the team discovered, depend on the activity of a gene called rutabaga, of which humans also have a similar version. A rapidly occurring, short-lived trace in a group of neurons that make up a structure called the “gamma” (γ) lobe produces a short-term memory. A slower, long-lived trace in the “alphabeta” (αβ) lobe fixes a long-term memory.
A tale of two lobes
Neuroscientists call the rutabaga gene a coincidence detector because it codes for an enzyme whose activity levels get a big boost when a fly perceives two stimuli that it has to learn to associate with one another. This enzymatic activity in turn signals to other genes critical for learning and memory.
A classic experiment that teaches flies to associate stimuli—and one that the CSHL team used—is to place them in a training tube attached to an electric grid, and to administer shocks through the grid right after a certain odor is piped into the tube. Flies with normal rutabaga genes learn to associate the odor with the shock and if given a choice, buzz away from the grid. But flies that carry a mutated version of rutabaga in their brains lack both short- and long-term memory, don’t learn the association, and so fail to avoid the shocks.
The team has now found, however, that this total memory deficit does not occur when flies carry the mutated version in either the γ or in the αβ lobes. Flies in which normal rutabaga function was restored within the γ lobe alone regained short-term memory but not long-term memory. Restoring the gene’s function in the αβ lobe alone restored long-term memory, but not short-term memory.
Long- and short-term memory involve different circuits
“This ability to independently restore either short- or long-term memory depending on where rutabaga is expressed supports the idea that there are different anatomical and circuit requirements for different stages of memory,” Dubnau explains. It also challenges a previously held notion that neurons that form short-term memory are also involved in storing long-term memory.
Previous biochemical studies have suggested that rapid, short-lived signals characteristic of short-term memory cause unstable changes in a neuron’s connectivity that are then stabilized by slower, long-lasting signals that help establish long-term memory in the same neuron. But anatomy studies have long hinted at different circuits. Surgical lesions that destroy different parts of an animal’s brain can separately disrupt the two kinds of memory, suggesting that the two memory types might involve different neuronal populations.
“We’ve now used genetics as a finer scalpel than surgery to reconcile these findings,” Dubnau says. His team’s results suggest that biochemical signaling for both types of memory are triggered at the same time, but in different neuron sets. Memory traces form more quickly in one set than the other, but the set that lags behind consolidates the memory and stores it long-term.
Why two mechanisms?
But why might the fly brain divide up the labor of storing different memory types this way? Dubnau’s hunch is that it might be because for every stimulus it receives, the brain creates its own representation of this information. And each time this stimulus—for example, an odor—is perceived again, the brain adds to the representation and modifies it. “Such modifications might eventually disrupt the brain’s ability to accurately remember that information,” Dubnau speculates. “It might be better to store long-term memories in a different place where there’s no such flux.”
The team’s next mission is to determine how much cross talk, if any, is required between the two lobes for long-term memory to get consolidated. This work will add to the progress that scientists have already made in treating memory deficits in humans with drugs aimed at molecular members of the rutabaga-signaling pathway to enhance its downstream effects.
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Citation
“Short- and Long-Term memory in Drosophila Require cAMP Signaling in Distinct Neuron Types,” appears in the August 25 issue of Current Biology. The full citation is: Allison. L. Blum, Wanhe Li, Mike Cressy, and Josh Dubnau. The paper is available online at http://www.cell.com/current-biology/abstract/S0960-9822(09)01398-0 (doi:10.1016/j.cub.2009.07.016)