Closely related organisms share most of their genes, but these similarities belie major differences in behavior, intelligence, and physical appearance.
Cold Spring Harbor, NY — Closely related organisms share most of their genes, but these similarities belie major differences in behavior, intelligence, and physical appearance. For example, we share nearly 99% of our genes with chimps, our closest relatives on the great “tree of life.” Still, the differences between the two species are unmistakable. If not just genes, what else accounts for the disparities? Scientists are beginning to appreciate that the timing of the events that happen during development plays a decisive role in defining an organism, which may help to explain how species evolve without the creation of new genes.
Today, a team of scientists at Cold Spring Harbor Laboratory (CSHL) has identified a key regulator of developmental timing. Led by CSHL Assistant Professor Christopher Hammell, the researchers describe how LIN-42, a gene that is found in animals across the evolutionary tree, governs a broad range of events throughout development.
“A great deal of science is focused on understanding how a single gene functions in the cell,” says Hammell. “But we are learning that when a gene is active is just as important as what it does.”
An organism develops in well-defined stages: nerves and muscles mature before reproductive tissues, for example. The stages unfold sequentially, like movements in a symphony. Played all at once, they would produce a terrible developmental cacophony, but with proper timing, a natural harmony can emerge.
Developmental stages are marked by the activation or repression of a specific and unique complement of genes, like individual notes within movements of a song. The order and duration of when these key developmental genes (or notes) are active (or played) within a given cell is controlled by a class of molecules called microRNAs (miRNAs). A single miRNA gene can control hundreds of other genes at once. If a miRNA turns off these specific genes too early or too late, the organism will suffer severe developmental defects. But little is known about how the activities of these miRNAs are regulated.
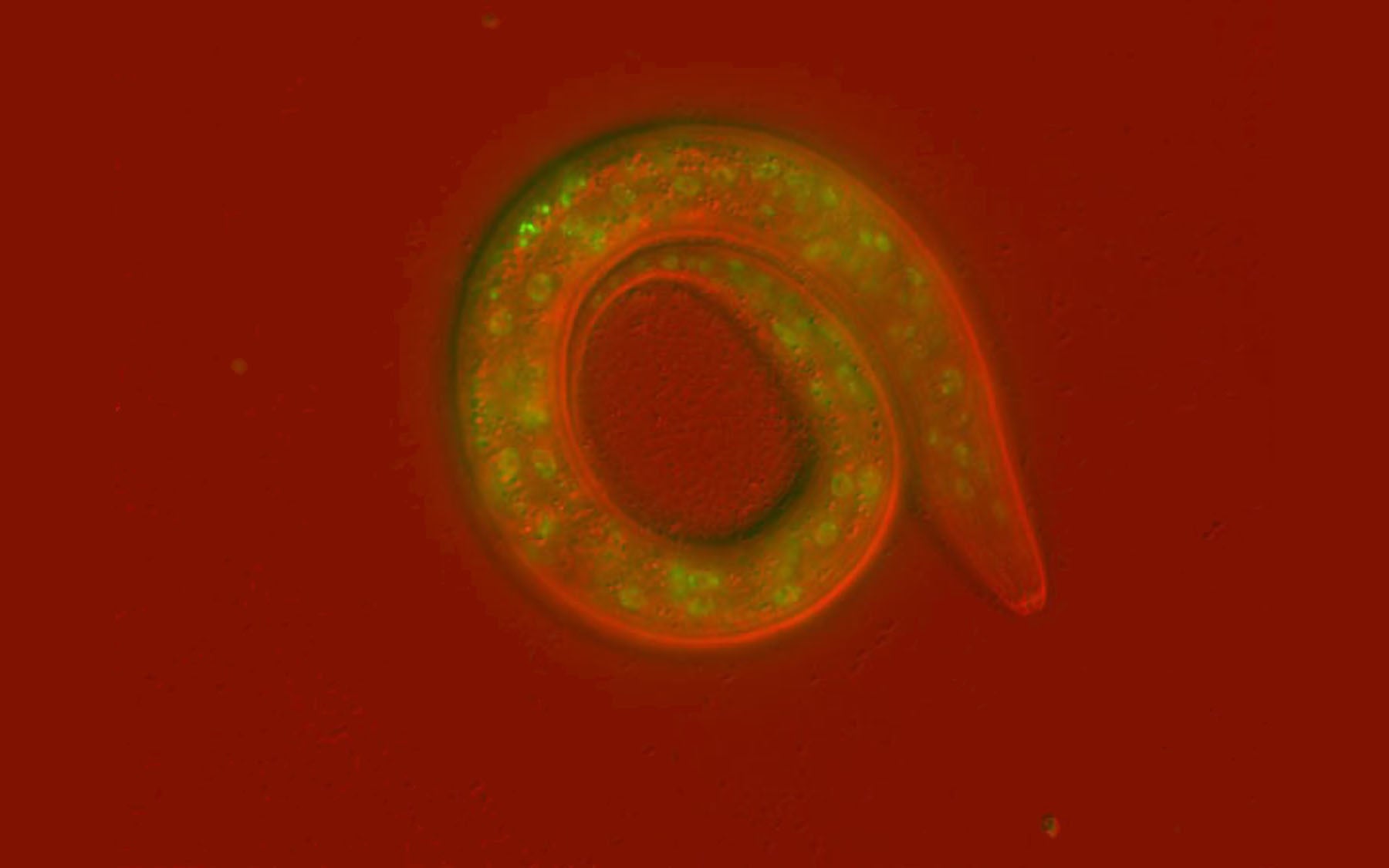
In work published today in PLoS Genetics, Hammell and his team describe the genetic approach they used to search for genes that control developmental timing through miRNAs. The team uses a tiny roundworm, called C. elegans, as a simple model for the events that occur during development, even in higher organisms. These worms have a fixed number of cells and each cell division is precisely timed. “It is the perfect model for our work,” says Hammell. “It enables us to understand exactly how a mutation affects development, whether maturation is precocious or delayed, by directly observing defects in the timing of gene expression.”
The team’s search uncovered the gene LIN-42 as a crucial regulator of developmental timing via its pervasive role in controlling miRNAs. “LIN-42 shares a significant amount of similarity to the genes that control circadian rhythms in organisms such as mice and humans,” says Roberto Perales, Ph.D., one of the lead authors of the study. “These are genes that control the timing of cellular processes on a daily basis for you and me. In the worm, these same genes and mechanisms control development, growth, and behavior and this system will provide us with leverage to understand how all of these things are coordinated.”
Hammell and his team found that LIN-42 controls the repression of numerous genes in addition to miRNAs. They also discovered that levels of the protein encoded by LIN-42 tend to oscillate over the course of development and form a part of a developmental clock. “LIN-42 provides the organism with a kind of cadence or temporal memory, so that it can remember that it has completed one developmental step before it moves on to the next,” says Hammell. “This way, LIN-42 coordinates optimal levels of the genes required throughout development.”
Written by: Jaclyn Jansen, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
This work was supported by the Rita Allen Foundation of which CMH is a Milton E. Cassel Scholar, the National Institutes of Health, and the Robertson Research Fund of Cold Spring Harbor Laboratory.
Citation
“LIN-42, the Caenorhabditis elegans PERIOD homolog, negatively regulates microRNA transcription” appears online in PLOS Genetics on July 17, 2014. The authors are: Roberto Perales, Dana King, Christina Aguirre-Chen, and Christopher Hammell. The paper can be obtained online at: http://www.plosgenetics.org
Principal Investigator

Christopher Hammell
Professor
Cancer Center Member
Ph.D., Dartmouth Medical School, 2002