If your DNA is a cookbook, a single gene is a recipe. But it’s a flexible recipe that if edited one way can make a pie; edited another way can make a cake. And a cake made the wrong way can mean cancer, as a team of researchers who looked at those gene editors writes in the recent issue of Cell Reports.
Those gene editors are known as splicing factors. When a gene is read out and copied, splicing factors choose where to cut and paste the copy so that it will give the right recipe to the cell for that moment in time.
“A gene can code for a protein that causes cell death, or a protein that prevents it, depending on the editing,” says breast cancer researcher Olga Anczukow, a molecular biologist who holds joint affiliations at the Jackson Laboratory for Genomic Medicine (JAX) and UConn Health. Anczukow was a former postdoc researcher in the lab of Cold Spring Harbor Laboratory Professor Adrian Krainer, a co-author on the paper.
Anczukow and colleagues at UConn Health, JAX and Cold Spring Harbor Laboratory were curious whether splicing factors could be responsible for the way some breast cancers grow and spread through the body. If a splicing factor was messing up, giving the cell the wrong recipe, it could cause the cell to behave badly and grow out of control, or migrate through the body and cause cancers elsewhere, what’s called metastasis.
They looked at cells from breast cancers and found that only a few splicing factors seemed connected to the cell’s cancerous behavior. In particular, three splicing factors gave the cells the same wrong recipe, enhancing the cell’s ability to grow and metastasize. Among these, a splicing factor called TRA2β seemed particularly enriched in triple negative breast cancers. Triple negatives are the worst breast cancers: They have the highest rates of metastasis, worst prognosis, and no targeted treatments.
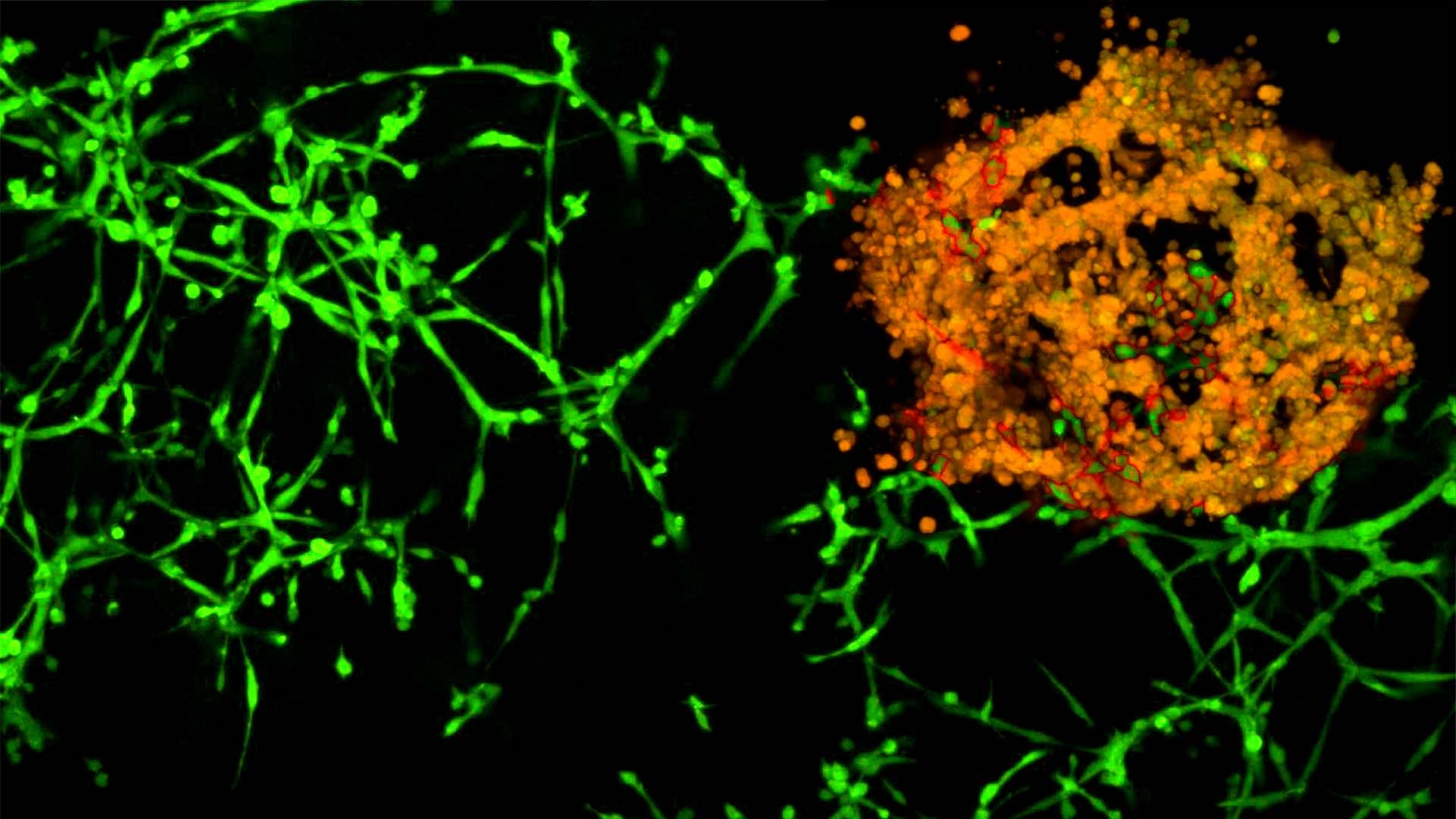
The researchers blocked TRA2β expression in cells, tiny tumors in a petri dish, and in mice. In all three situations, cells lacking TRA2β were unable to metastasize.
Identifying TRA2β was exciting, the researchers said. Finding a way to block it could provide a treatment for this most dreaded form of breast cancer. They hope to learn more about how the splicing factors become dysregulated, and eventually develop a drug to target them.
Targeting splicing defects has become a reality with the approval of Spinraza®, a first of its kind drug that corrects abnormal splicing. Spinraza® was developed by Krainer and is the first FDA-approved drug to treat children with spinal muscular atrophy. Researchers hope that in the future this type of drug can be used to treat other diseases with splicing defects, including cancer.
“Once we know that a splicing factor promotes disease, a targeted treatment could involve a selective inhibitor of that factor, or by restoring the correct editing of one or more of its crucial targets,” Krainer said.
Written by: Kim Krieger, UConn Communications
Funding
The research was funded by the National Cancer Institute and National Institute of General Medical Sciences at the National Institutes of Health, the Susan G. Komen Foundation for the Cure, the Terri Brodeur Breast Cancer Foundation, and JAX start-up funds.