p53 gene network is key determinant of response; its attenuation causes drug resistance
Cold Spring Harbor, NY — Being able to accurately predict how a given cancer will respond to chemotherapy would spare patients with non-responsive tumors the burden of dealing with a toxic and ultimately unhelpful treatment. Just as important, knowing which of a patient’s cancer-causing genetic lesions are contributing to drug resistance might help doctors redesign therapy for maximum benefit.

Researchers led by Professor Scott Lowe, Ph.D., of Cold Spring Harbor Laboratory (CSHL), have come closer to achieving these critical goals for human cancer therapy by developing new mouse models for human acute myeloid leukemia (AML), an aggressive and devastating cancer of white blood cells.
As Lowe and colleagues report in the April 1st issue of the journal Genes and Development, their models, which include treatment protocols that closely mimic current AML therapy in people, precisely recapitulate genetic associations that have been linked to favorable or adverse treatment responses in patients. These findings provide compelling evidence for the notion that such models can predict how human cancers will respond to therapy and help to identify genes promoting resistance or sensitivity to any cancer drug. The mouse models, the CSHL team notes, also serve as an effective test system for new drugs and treatment strategies in AML.
Need for a reliable prediction model
“By giving us a better understanding of how cancer’s genetic makeup, or ‘genotype,’ influences the outcomes of treatment, these models might help improve how existing drugs are used in people, and spur the design of more effective therapies,” Lowe observes. “The new models are an important preclinical tool that will allow knowledge gained from cancer genetics to be put to effective use in the clinic.”
The Lowe team’s mouse models also provide valuable insights into how leukemia develops and progresses. Their study traces the intracellular network controlled by the p53 gene, a linchpin of the cell’s anti-tumor defense response, as the key determinant of AML’s aggressiveness and response to therapy.
Most patients with AML receive the same standardized treatment—an initial phase of intense chemotherapy followed by additional chemotherapy cycles or bone marrow transplantation. Yet only a quarter of patients are cured and most die within a few months. This diversity in treatment response is due to AML’s genetic heterogeneity, meaning that the hundred or so mutations associated with this form of cancer occur in different combinations in each patient and influence therapeutic outcomes in different ways.
Some gene mutations in cancer have been correlated with clinical outcome. But AML has proved to be genetically too complex, and the current experimental systems for predicting treatment response too unreliable for the information to be used in a standard way in the clinic.
These standard systems, in which anticancer drugs were tested in human cancer-cell lines, do not factor in the effects of a real tumor’s environment on its growth. At the same time, placing these cells into animals to create a so-called “xenotransplant” model has not been an effective solution, according to Johannes Zuber, M.D., a Clinical Fellow in the Lowe lab who played a key role in the research and is the first author on the team’s paper. The reason, explains Zuber, is that “the cells are so poorly defined at the genetic level and tend to defy analysis of the molecular factors that influence drug response.”
Making mice with human-like AML
Lowe’s group surmounted these problems by first identifying the most commonly occurring mutations in a group of 111 children with AML and then engineering these mutations into mice, which soon developed leukemia. Among the participating AML patients, the two most common mutations were observed to occur when chromosomes broke apart and reattached in new places. This incorrect fusion is thought to create cancer-causing genes, or “oncogenes,” that encode so-called fusion proteins, which profoundly disturb the developmental program of cells.
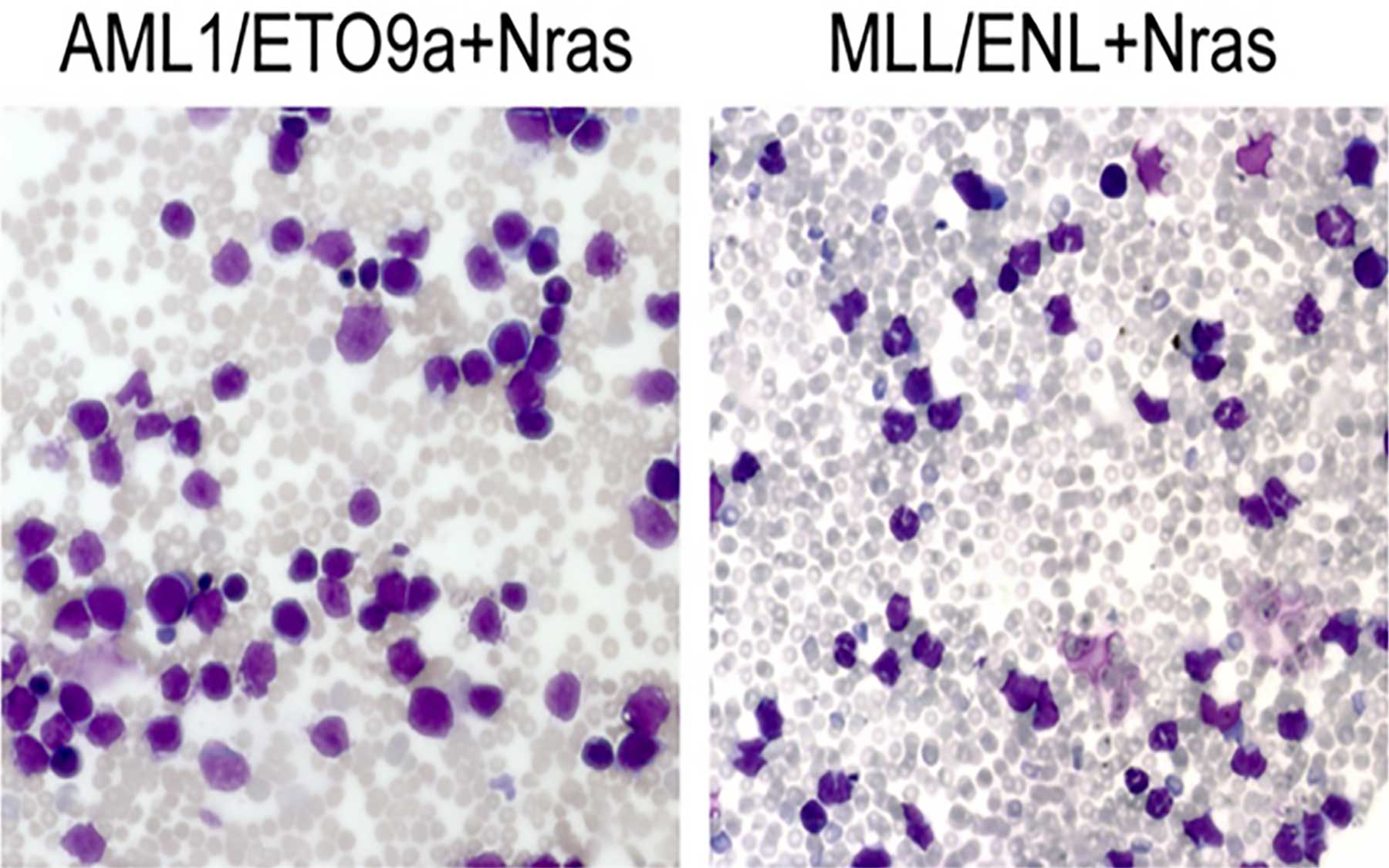
Both oncogenic mutations identified by Lowe’s team in AML patients have been well studied. The oncogene AML1/ETO previously has been associated with a favorable therapeutic outcome in people, while fusion oncogenes involving the MLL gene have been associated with an adverse outcome. To design an animal model that predicts these outcomes, the team introduced each mutation individually into stem and progenitor cells along with another oncogene, called Nras, which also appears frequently in human AML and is commonly found in concert with AML1/ETO and MLL fusion genes.
These altered stem cells were transplanted into mice that were first treated with radiation to destroy existing bone marrow cells. The altered stem cells then took over the “host” bone marrow and promoted the development of leukemia. To track leukemia onset and response to therapy, the scientists tagged all leukemia cells with a gene from fireflies that encodes an enzyme called luciferase, which makes leukemia cells glow and detectable by an ultrasensitive camera. Within weeks, the mice developed leukemias that showed the same genetic and pathological features as human AML.
“We were therefore certain,” says Lowe, “that these animals were good models for studying the impact of cancer’s genetic heterogeneity upon therapy response and getting a correct read-out in terms of outcome prediction.”
Sensitivity vs. resistance to chemotherapy
Experiments in which the team treated the mice with the same chemotherapy regimen administered to AML patients have proved Lowe to be correct. Just as in humans, leukemias in mice that received the AML1/ETO oncogene were also sensitive to chemotherapy and soon regressed, whereas MLL-triggered leukemias remained resistant and eventually killed their hosts.
The models also proved useful in helping the scientists identify the molecular basis of this difference in outcome. The chemosensitive mice with AML1/ETO-triggered leukemia had a distinct gene expression signature that was absent in the others; the genes whose expression was most significantly altered in the sensitive mice belong to a network controlled by p53—the gene that masterminds the cell’s anti-tumor defense.
Unlike the treatment-resistant mice, the treatment-sensitive mice showed a robust activation of the p53 network. This activation was critical for a favorable outcome: when the team knocked out p53 in the chemosensitive mice, their leukemia became drug-resistant and aggressive.
“These results in the new animal models suggest that AML patients diagnosed with the MLL chromosome rearrangement will not show durable responses to standard chemotherapy,” according to Lowe, Zuber, and colleagues. “These patients should be offered alternative therapeutic approaches at diagnosis.”
In addition to Lowe’s group at CSHL, the study also includes researchers from St. Jude’s Hospital, Tennessee; Stony Brook University, New York; and the Universities of Chicago and San Francisco.
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Citation
“Mouse models of human AML accurately predict chemotherapy response,” appears in the April 1st issue of Genes and Development. The full citation is: Johannes Zuber, Ina Radtke, Timothy S. Pardee, Zhen Zhao, Amy Rappaport, Weijun Luo, Mila E. McCurrach, Miao-Miao Yang, M. Eileen Dolan, Scott C. Kogan, James R. Downing, and Scott W. Lowe. The article can be found online at http://genesdev.cshlp.org/