CSHL Professor Alea Mills explains what’s so important about the Chd5 tumor suppressor and what her team has discovered about it.
Cold Spring Harbor, NY — A team of cancer researchers at Cold Spring Harbor Laboratory (CSHL) has solved the mystery of how one of the most powerful of the body’s natural tumor-suppressing proteins, called Chd5, exerts its beneficial effects.
The findings, published online today in the journal Cell Reports, are important because Chd5 engages processes fundamental to cancer prevention. Conversely, when Chd5 is mutated or missing, an important door is opened to cancer initiation.
“For this reason, figuring out the mechanics of how Chd5 works to prevent cancer can directly impact the treatment of a diverse array of human cancers,” says Alea A. Mills, Ph.D., team leader and Professor at CSHL. “Until now, we didn’t understand very much about this process.”
Since the time that Mills first discovered Chd5 as a tumor suppressor in 2007 and reported that it facilitates the expression of a tumor-suppressive network—acting as a “master switch” for several tumor suppressor proteins—Chd5 has been shown to predict survival following anticancer therapy. That is, patients struck with malignancies such as neuroblastoma, gall bladder cancer, and ovarian cancer live much longer if they have high levels of Chd5.
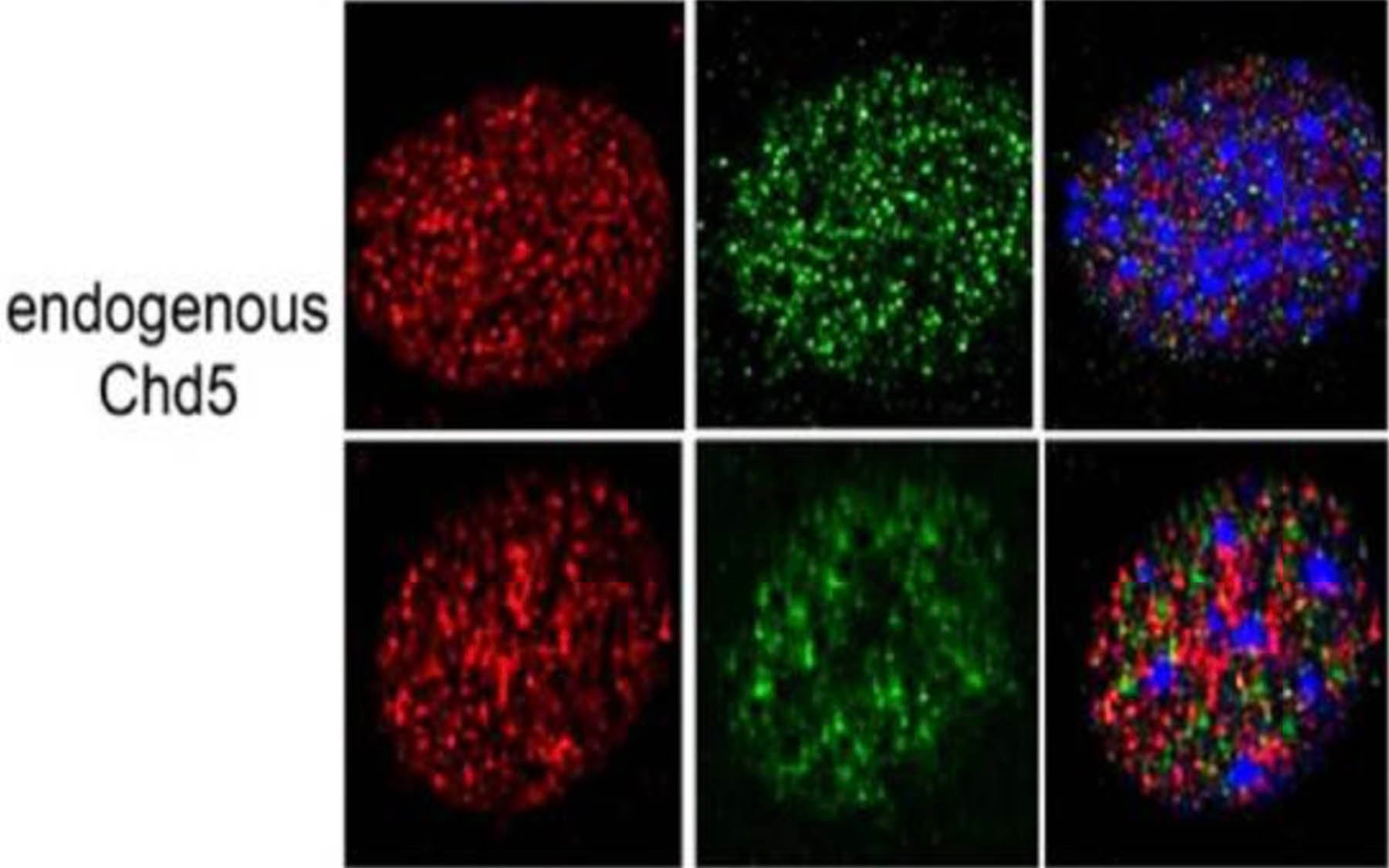
Now, Mills’ team has discovered that Chd5’s beneficial activity occurs when the protein binds to another protein called histone H3, which helps bundle the genetic material in the nucleus of cells.
Her team began their work knowing that Chd5 was a member of a family of proteins having at least nine members. Called chromatin remodeling proteins, they are involved in the immensely important job of shaping and chemically marking the highly compressed bundles of DNA and protein that biologists call chromatin.
Among other things, the degree to which chromatin is compressed helps determine whether particular genes among our total complement of 21,000 are either accessible or inaccessible to molecular machines that help to “express” them. Broadly speaking, tightly bundled chromatin is inaccessible to the gene-expression machinery, causing genes located in a given bundle to be shut off. The reverse is true of loosely packed chromatin.
The CSHL team’s new finding involves Chd5’s role in another process through which chromatin can be shaped and modified. Namely, via reading molecular groups on chromatin that effectively flag parts of its structure, a process called epigenetic marking.
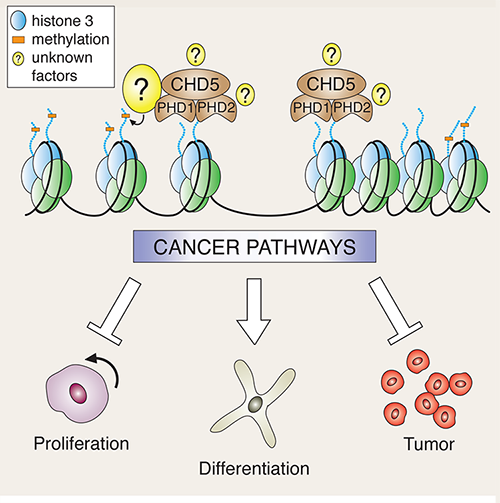
Mills explains: “Our new work demonstrates that the Chd5 protein can activate the tumor-suppressive network when a portion of its structure (called PHD domains) bind to histone H3, one of the five histone proteins that make up the main structure of chromatin. Importantly, we found that this binding occurs specifically when the tail-like portion of histone H3 is what we call ‘naked’—that is, free of any epigenetic marks.”
The team showed in living mice that when the gene that expresses Chd5 is mutated, yielding a protein that cannot bind the H3 histone, Chd5’s usual tumor-suppressive effects do not occur; it no longer inhibits proliferation, thereby setting the stage for cancer initiation. The cancer used as a model for this part of the work was human neuroblastoma.
The team reported other details of the mechanism behind Chd5’s tumor suppression, revealing how the interaction of Chd5 and naked H3 histones orchestrates programs that prevent cancer, mainly by binding to genes and affecting their “on” vs. “off” state.
“It is likely that Chd5’s multi-faceted ways of enforcing tumor suppression will prove useful for regulating diverse types of cancers, including cancers with many different combinations of genetic lesions,” Mills says.
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
This work was supported by The SASS Foundation for Medical Research and the National Cancer Institute [NIH RO1 CA127383].
Citation
“Chd5 Requires PHD-mediated Histone 3 binding for Tumor Suppression” appears online Thursday, January 10, 2013 in Cell Reports. The authors are: Shilpi Paul, Alex Kuo, Thomas Schalch, Hannes Vogel, Leemor Joshua-Tor, W. Richard McCombie, Or Gozani, Molly Hammell and Alea A. Mills. The paper can be obtained at: http://cellreports.cell.com/
Principal Investigator

Alea A. Mills
Professor
Cancer Center Member
Ph.D., University of California, Irvine, 1997