CSHL’s Rob Martienssen and W. Richard McCombie help to develop a genome “tree of life,” the largest yet for seed plants
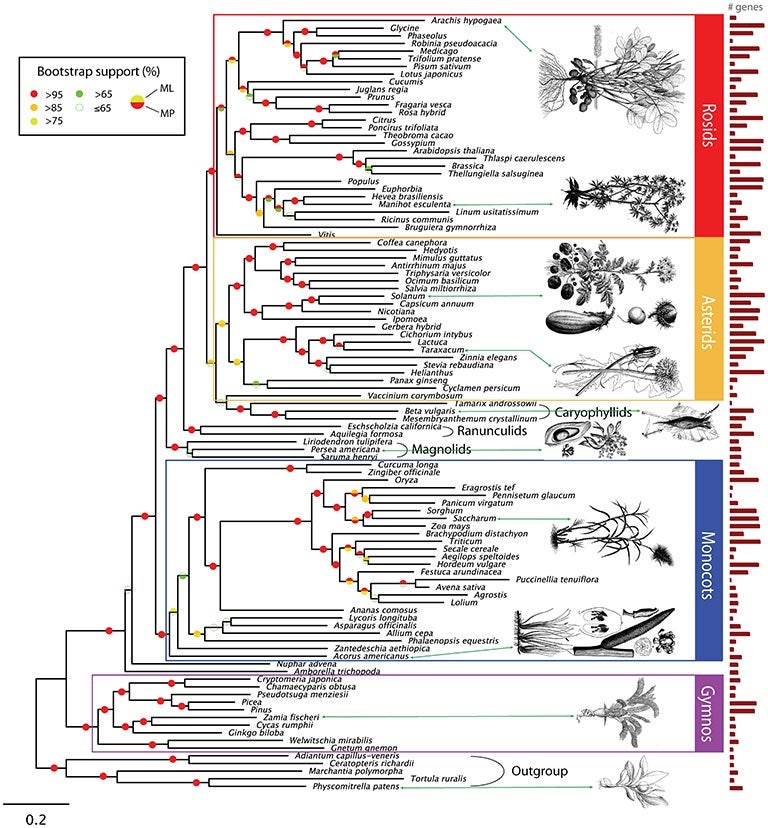
Cold Spring Harbor, NY — Scientists at Cold Spring Harbor Laboratory, the American Museum of Natural History, the New York Botanical Garden, and New York University have created the largest genome-based “tree of life” for seed plants to date. Their findings, published today in the journal PLoS Genetics, plot the evolutionary relationships of 150 species of plants based on advanced genome-wide analysis of gene structure and function. This new approach, called functional phylogenomics, allows scientists to reconstruct the pattern of events that led to the vast number of plant species we see today, and could help identify genes that can be used to improve seed quality for agricultural and environmental purposes.
“Ever since Darwin first described the ‘abominable mystery’ behind the rapid explosion of flowering plants in the fossil record, evolutionary biologists have been trying to understand the genetic and genomic basis of the astounding diversity of plant species,” said Rob DeSalle, corresponding author on the paper and a curator in the Museum’s Division of Invertebrate Zoology and the Sackler Institute for Comparative Genomics. “Having the architecture of this plant tree of life allows us to start to decipher some of the interesting aspects of evolutionary innovations that have occurred in this group.”
The research, performed by members of the New York Plant Genomics Consortium, was funded by the National Science Foundation (NSF) Plant Genome Program to identify the genes that caused the evolution of seeds, an important economic trait. The group selected 150 representative species from all of the major seed plant groups. The species span from the flowering variety—peanut and dandelions, for example—to non-flowering cone plants like spruce and pine. The sequences of the plants’ genomes—all of the biological information needed to build and maintain an organism, encoded in DNA—were either culled from pre-existing databases or generated in the field and at the New York Botanical Garden in the Bronx, from live specimens.
With new algorithms developed at the Museum and NYU and the processing power of supercomputers at Cold Spring Harbor Laboratory and overseas, the sequences—nearly 23,000 sets of genes (specific sections of DNA that code for certain proteins)—were grouped, ordered, and placed in an organizational tree according to their evolutionary relationships. In addition, algorithms determining similarities of biological processes were used to identify the genes underlying species diversity.
“Previously, phylogenetic trees were constructed from standard sets of genes and were used to identify the relationships of species,” said Gloria Coruzzi, a professor in New York University’s Center for Genomics and Systems Biology and the principal investigator of the NSF grant. “In our novel approach, we create the phylogeny based on all the genes in a genome, and then use the phylogeny to identify which genes provide positive support for the divergence of species.”
The researchers’ results support major hypotheses about evolutionary relationships in seed plants. The most interesting finding is that gnetophytes, a group that consists mostly of shrubs and woody vines, are the most primitive living non-flowering seed plants—present since the Mesozoic era, the “age of dinosaurs.” They are situated at the base of the evolutionary tree of seed plants.
“This study resolves the longstanding problem of producing an unequivocal evolutionary tree of the seed plants,” said Dennis Stevenson, vice president for laboratory research at the New York Botanical Garden. “We can use this information to determine when and where important adaptations occur and how they relate to plant diversification.”
The researchers were also able to make predictions about genes that caused the evolution of important plant characteristics. One such evolutionary signal is RNA interference, a process that cells use to turn down or silence the activity of specific genes. Based on their new phylogenomic maps, the researchers believe that RNA interference played a large role in the separation of monocots—plants that have a single seed leaf, including orchids, rice, and sugar cane—from other flowering plants.
“Even more surprising, RNA interference also played a major role in the emergence of flowering plants themselves,” said Rob Martienssen, Professor and HHMI-GBMF Investigator at Cold Spring Harbor Laboratory. “Genes required for the production of small RNA in seeds were at the very top of the list of genes responsible for the evolution of flowering plants from cone-bearing plants. In collaboration with colleagues in Mexico last year, we found that these same genes control various aspects of maternal reproduction, providing remarkable insight into the evolution of reproductive strategy in flowering plants,” he explained.
The data and software resources generated by the researchers are publicly available and will empower other comparative genomic researchers to exploit plant diversity to identify genes associated with a trait of interest or agronomic value. These studies could have implications for improving the quality of seeds, which are the basis of much of today’s agriculture products, ranging from food to clothing.
The phylogenomic approach used in this study also could be applied to other groups of organisms to further explore how species originated, expanded, and diversified.
“The collaboration among the institutions involved here is a great example of how modern science works,” said Sergios-Orestis Kolokotronis, a term assistant professor at Columbia University’s Barnard College and a research associate at the Museum’s Sackler Institute. “Each of the four institutions involved has its own strengths and these strengths were nicely interwoven to produce a novel vision of plant evolution.”
Written by: Hema Bashyam, Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
The New York Plant Genomics Consortium is supported by funding from funding from the U.S. National Science Foundation.
Citation
“A Functional Phylogenomic View of the Seed Plants,” appears in PLoS Genetics online at 5pm on December 15. The full citation is Ernest K. Lee, Angelica Cibrian-Jaramillo, Sergios-Orestis Kolokotronis, Manpreet S. Katari, Alexandros Stamatakis, Michael Ott, Joanna C. Chiu, Damon P. Little, Dennis Wm. Stevenson, W. Richard McCombie, Robert A. Martienssen, Gloria Coruzzi, Rob DeSalle. The paper can be downloaded at http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1002411