Scientists at Cold Spring Harbor Laboratory (CSHL) have devised a powerful algorithm that improves the effectiveness of an important research technology harnessing RNA interference, or RNAi.
Cold Spring Harbor, NY — Scientists at Cold Spring Harbor Laboratory (CSHL) have devised a powerful algorithm that improves the effectiveness of an important research technology harnessing RNA interference, or RNAi.
Discovered in the late 1990s, RNAi is a naturally occurring biological mechanism in which short RNA molecules bind to and “interfere” with messages sent by genes that contain instructions for protein production. Such interference can prevent a gene from being expressed. In addition to helping regulate gene expression, the RNAi pathway in many species, including humans, acts to defend the genome from parasitic viruses and transposons.
Harnessed by scientists since the mid-2000s, RNAi has provided a way to artificially “knock down” the expression of specific genes. By preventing a gene or genes from being activated in a model organism such as a mouse, for instance, much can be learned by inference about gene function. RNAi-based technology also has been extremely useful as tool in drug discovery.
At CSHL, HHMI Professor Gregory Hannon and colleagues have made seminal discoveries over the last decade in elucidating the mechanism through which RNAi operates in nature. They have also pioneered means of harnessing the RNAi pathway, creating comprehensive libraries of small RNA molecules—called short hairpin RNAs, or shRNAs. These have enabled researchers worldwide to selectively shut down specific genes, for instance, across the entire human and mouse genomes.
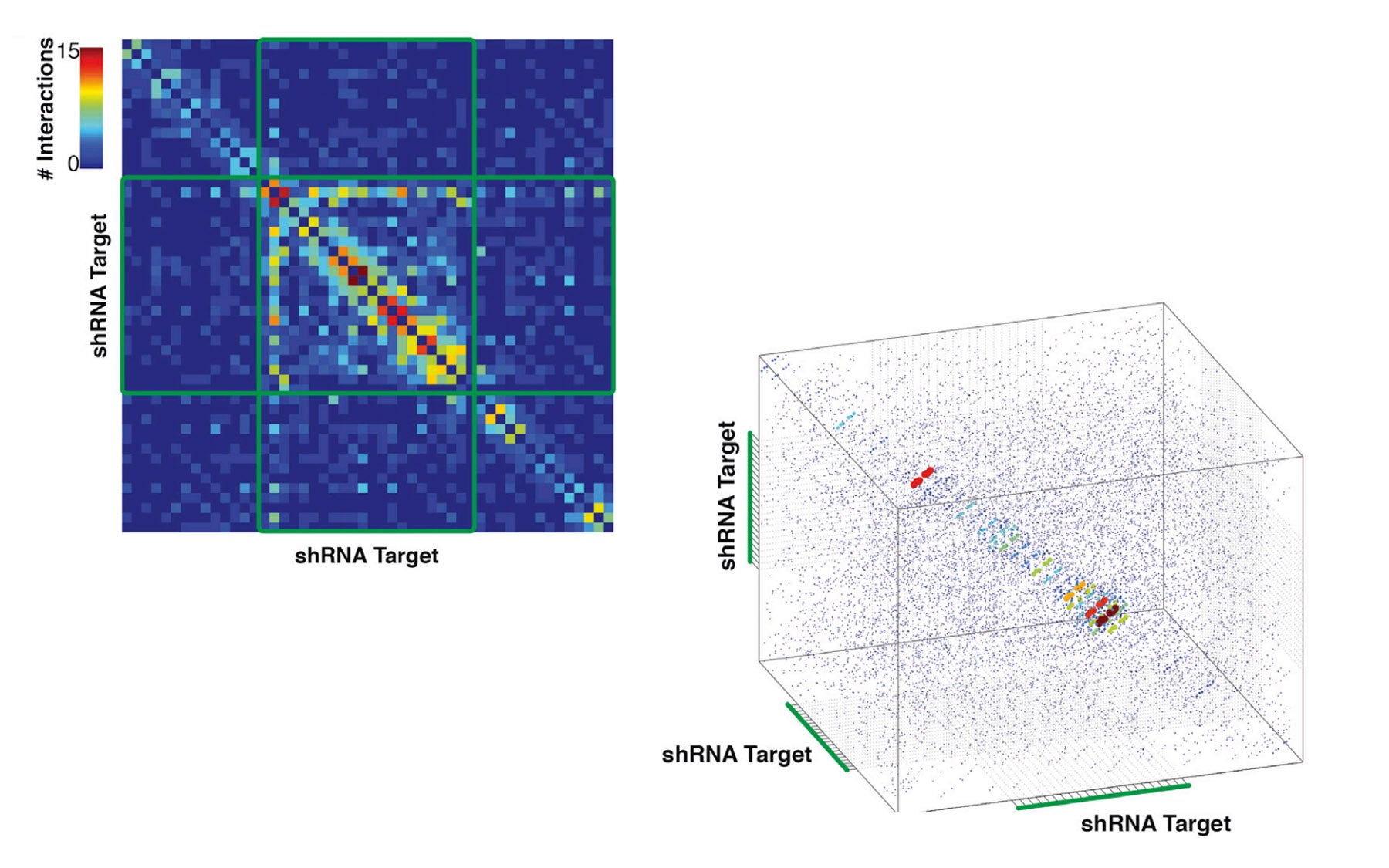
In research published ahead of print November 26 in Molecular Cell, a Hannon lab team led by Research Investigator Simon Knott reported the creation of an algorithm that enables researchers to identify shRNAs that will be most effective for shutting down specific genes. Before this work, the sequence determinants responsible for optimizing shRNA efficacy were largely unknown. Thus the new research addresses an important technical issue in using RNAi for gene knockdown. The new algorithm was trained to identify sequence determinants and is able to search across each gene and distinguish the sequence targets that will produce the most effective gene knockdown.
Development of the algorithm, called shERWOOD, was based on a massive parallel assessment of the potency of some 250,000 different shRNAs. In effect, a computer was trained to make very accurate predictions, by learning what distinguished the potent shRNAs within this set from those that were less potent. After this learning process the computer was then able to predict the potency of new sequences.
“We built upon this result to design and construct next-generation shRNA libraries targeting the exomes, or protein-encoding gene set, of mice and humans,” says Dr. Knott.
shERWOOD-based libraries are being made available to the research community by Transomic Technologies (Huntsville, Alabama).
Written by: Peter Tarr, Senior Science Writer | publicaffairs@cshl.edu | 516-367-8455
Funding
The research described in this release was supported by the Howard Hughes Medical Institute, the National Institutes of Health, Kathryn W. Davis, the Hope Funds for Cancer Research and the Boehringer Ingelheim Foundation.
Citation
“A Computational Algorithm to Predict shRNA Potency” appeared November 26, 2014 in Molecular Cell. The authors are: Simon R.V. Knott, Ashley R. Maceli, Nicolas Erard, Kenneth Chang, Krista Marran, Xin Zhou, Assaf Gordon, Osama El Demerdash, Elvin Wagenblast, Sun Kim, Christof Fellmann and Gregory J. Hannon. The paper can be obtained at: http://www.sciencedirect.com/science/article/pii/S1097276514008351