Plant genomics has come a long way since Cold Spring Harbor Laboratory (CSHL) helped sequence the first plant genome. But engineering the perfect crop is still, in many ways, a game of chance. Making the same DNA mutation in two different plants doesn’t always give us the crop traits we want. The question is why not? CSHL plant biologists just dug up a reason.
CSHL Professor and HHMI Investigator Zachary Lippman and his team discovered that tomato and Arabidopsis thaliana plants can use very different regulatory systems to control the same exact gene. Incredibly, they linked this behavior to extreme genetic makeovers that occurred over 125 million years of evolution.
The scientists used genome editing to create over 70 mutant strains of tomato and Arabidopsis thaliana plants. Each mutation deleted a piece of regulatory DNA around a gene known as CLV3. They then analyzed the effect each mutation had on plant growth and development. When the DNA keeping CLV3 in check was mutated too much, fruit growth exploded. Danielle Ciren, a recent CSHL School of Biological Sciences graduate who led this study, explains:
“CLV3 helps plants develop normally. If it wasn’t turned on at the exact time that it is, then plants would look very different. All the fruits would be ginormous and not ideal. You have to balance growth and yield. If a plant has giant tomatoes but only two, is that as beneficial as a lower yield? There’s no simple solution. You’re always sacrificing something when you’re trying to get something improved.”
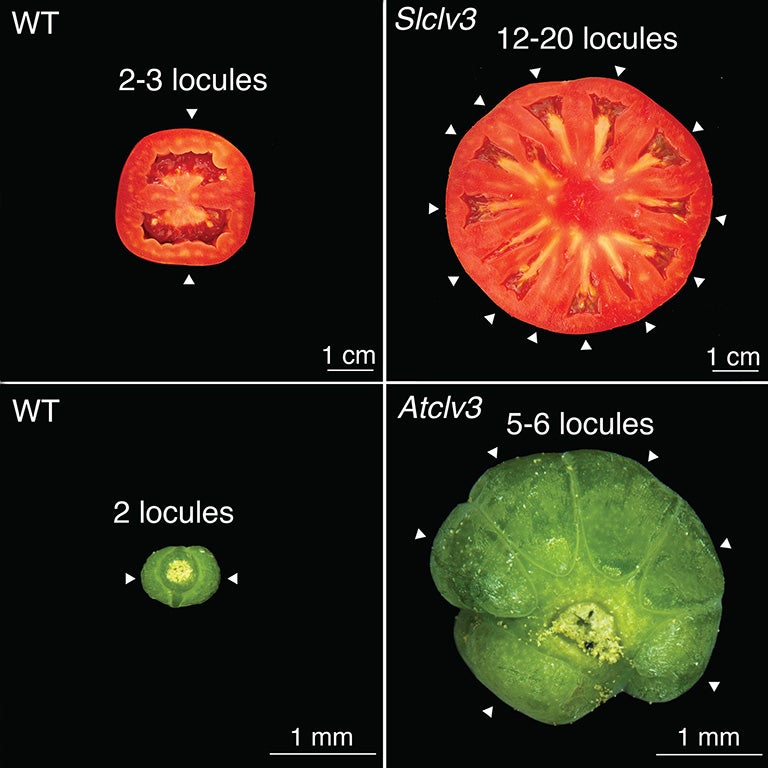
For tomatoes, engineering mutations near the beginning but not the end of the CLV3 gene dramatically affected fruit size. For Arabidopsis, areas around both parts of the gene needed to be disrupted. This indicates something happened over the last 125 million years that made the plants evolve differently. Exactly what occurred remains a mystery. Ciren explains:
“You can’t go back to the common ancestor because they don’t exist anymore. So it’s hard to say what was the original state and how have things been mixed up. The most simple explanation is that there’s a regulatory element that’s conserved in some capacity, and it’s been altered in subtle ways. It is a bit unexpected.”
What is certain is that genetic regulation is not uniform between plant species. Unearthing these genetic differences could help make crop genome engineering more predictable. And that would be a big win not just for science but for farmers and plant breeders across the globe.
Written by: Luis Sandoval, Communications Specialist | sandova@cshl.edu | 516-367-6826
Funding
Natural Sciences and Engineering Research Council of Canada, National Institute of General Medical Sciences, Howard Hughes Medical Institute, National Science Foundation Plant Genome Research Program
Citation
Ciren, D., et al., “Extreme restructuring of cis-regulatory regions controlling a deeply conserved plant stem cell regulator”, PLOS Genetics, March 4, 2024. DOI: 10.1371/journal.pgen.1011174
Principal Investigator

Zachary Lippman
Professor & HHMI Investigator
Jacob Goldfield Professor of Genetics
Director of Graduate Studies
Ph.D., Watson School of Biological Sciences at Cold Spring Harbor Laboratory, 2004
