Tracking a person entails searching through their email, phone, and other means of communication to map out their network. To do this for a brain cell, more creativity is called for.
After more than a century of investigation into the diverse cells of the brain, neuroscientists still are not sure what exactly makes one neuron different from another. They know how certain brain cells look, whether they inhibit or excite, and when they are most active, but these measures can’t unambiguously identify a neuron, just as appearance or outward behavior doesn’t capture the identity of a person.“What the intelligence community does to track someone is find out who you are talking to and using what medium,” explains neuroscientist Anirban Paul. This approach is incredibly powerful for identifying brain cells, too, he and colleagues including Professor Josh Huang reported in a recent paper. For a person, it entails searching through your email, your phone, and other means of communication to map out your network. To do this for a brain cell, more creativity is called for.
So far, a common approach to the brain cell identity problem has been to categorize the cells based on certain “marker” molecules they display, which Paul finds as unsatisfying as categorizing people based on their accessories. If you “imagine 10 photos of me, but in each one I’m wearing a different hat,” he asks, “are these different people, or do they just look different?” Like hats, markers on cells can easily serve as a disguise, resulting in categories that don’t mean very much, he says.
That’s why Paul sought a way to let the brain cells tell him what makes them unique. To do that, he focused on molecules called mRNAs, which deliver messages from a cell’s DNA to the molecular machinery that carries out its genetic instructions. Each mRNA corresponds to a particular gene. Instead of single marker mRNAs, he used mRNAs from more than 600 gene “families,” in which members play related molecular roles.
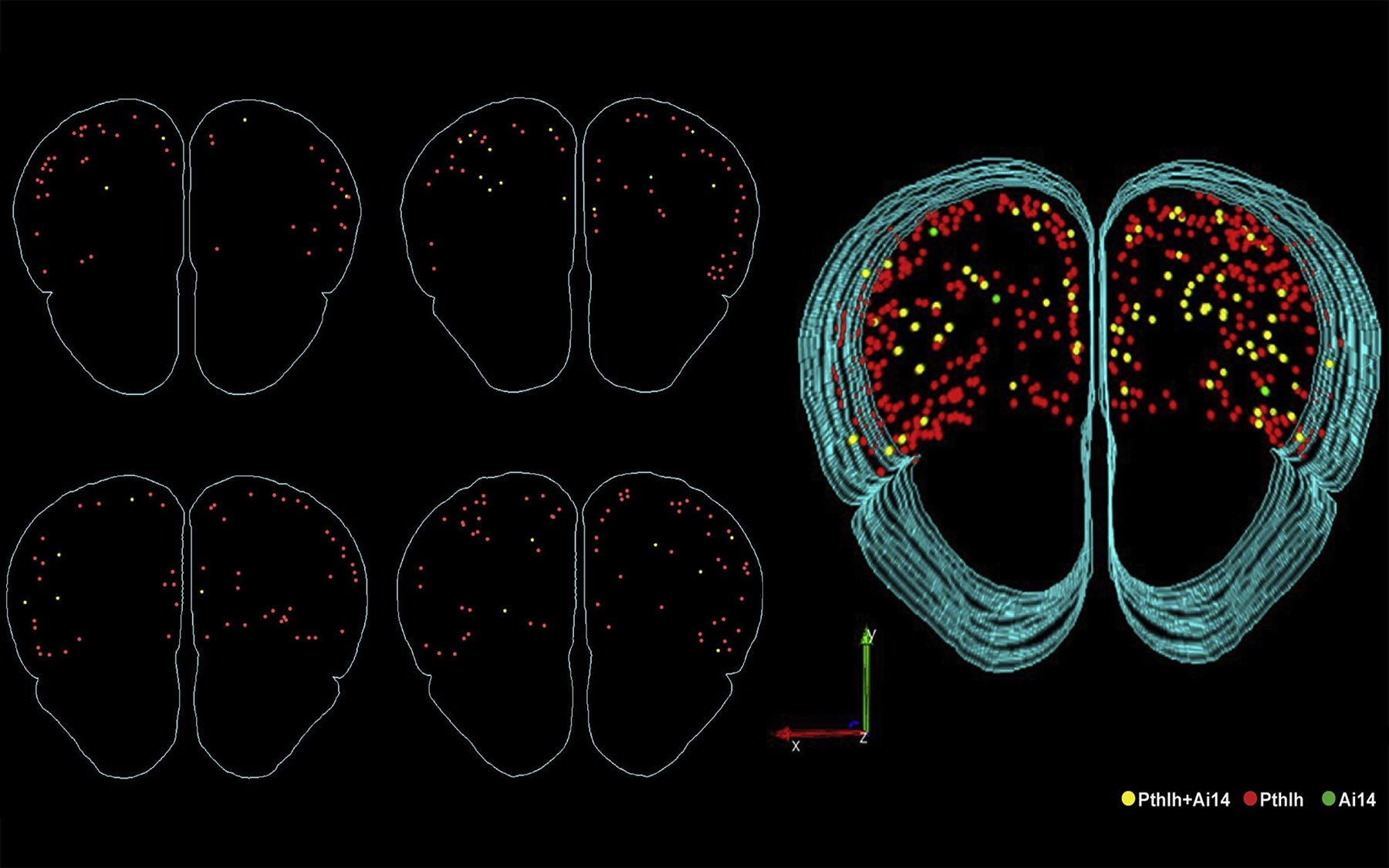
He started with six brain cell subtypes that most neuroscientists already agree are distinct, because, besides extensive past studies, “just by looking at them, you know they’re different.” From those neurons, he collected mRNA data for all 20,000 human gene equivalents in mouse and asked which families are actually the most different in these six types.
This spreadsheet full of information on the mRNA molecules floating around each brain cell was kind of like all of the digital information that a CIA agent would get from someone’s smart phone—their browsing history, the apps they use, their emails, and so on. The agent would know to look through the person’s texts and emails and calls to figure out whether the person is truly a member of, say, the Mafia. But Paul didn’t have the advantage of knowing not to bother with the cellular equivalent of a person’s Candy Crush scores. And the dataset he had was enormous.
To start making sense of the millions upon millions of data points on the cells’ mRNAs, each corresponding to the activity of a certain gene, Paul sat down each morning with the leader of his lab, Professor Josh Huang. They broke the data up into lists of different kinds of genes and ran these chunks of data through a computer program to see if that information was good enough to tell apart two neuron types that they were already knew were different. This seemingly dull task became a “kind of high we needed every morning,” he says, because a picture of what reliably determines the identity of a brain cell began to emerge bit by bit.
As their program told them which genes were proving useful in characterizing a cell, they went to the whiteboard to chart the location of the protein each gene produces. Amazingly, they all mapped to the interface between neurons, called the synapse, which is where neurons send messages to each other. Each morning, the pair would continue to find that the identifying genes “all point to the single quintessential function of cell to cell communication,” says Paul.
It was so exhilarating that he and Huang would take photos of their whiteboard sketches with their phones and send them to each other even when they were apart, because they couldn’t wait to show each other. All of the “identifying” genes encoded proteins that are involved in enabling neurons to communicate. In a sense, neurons “are” who they communicate with—and, by the same token, who they cannot speak with, because of incompatible communication equipment.
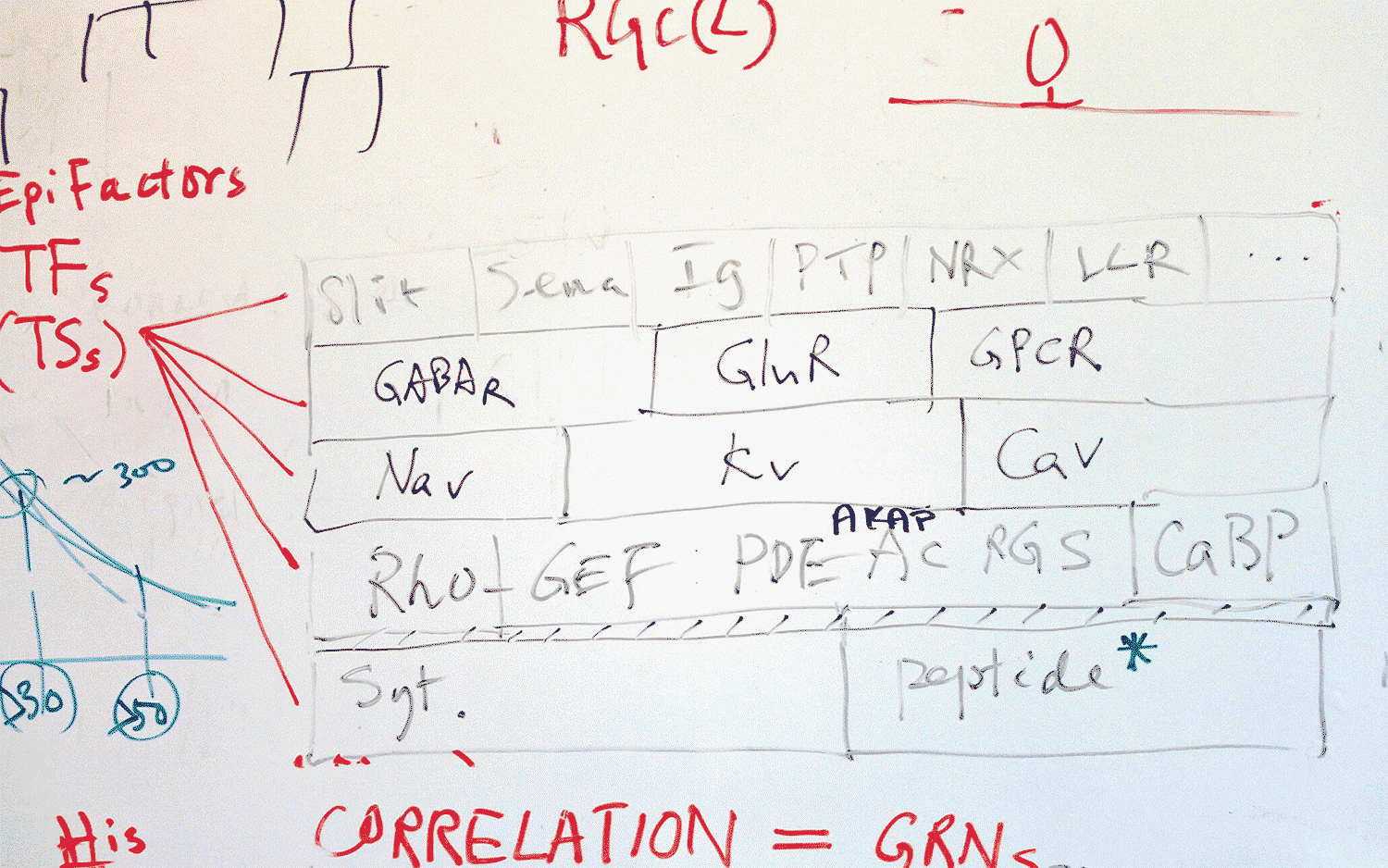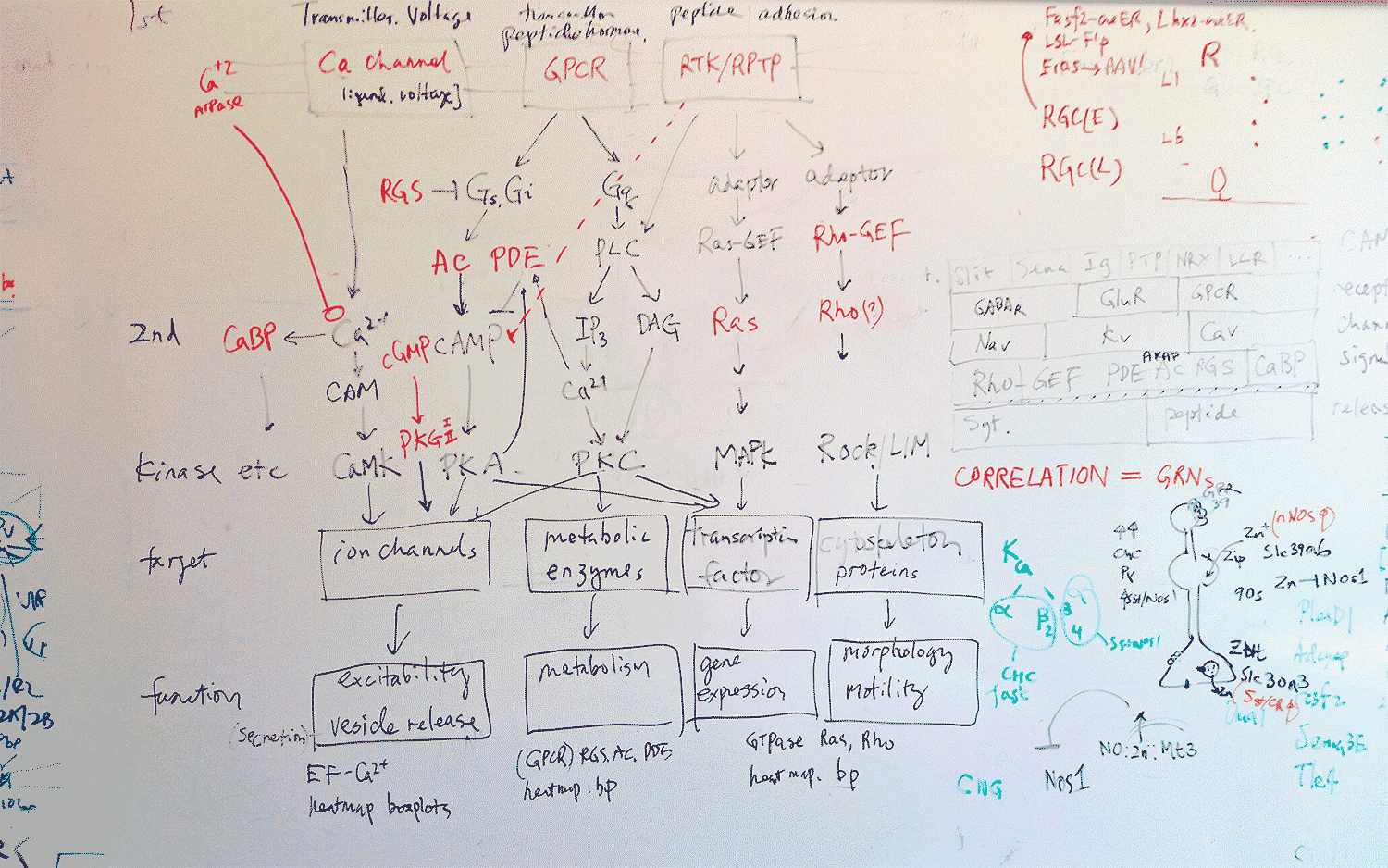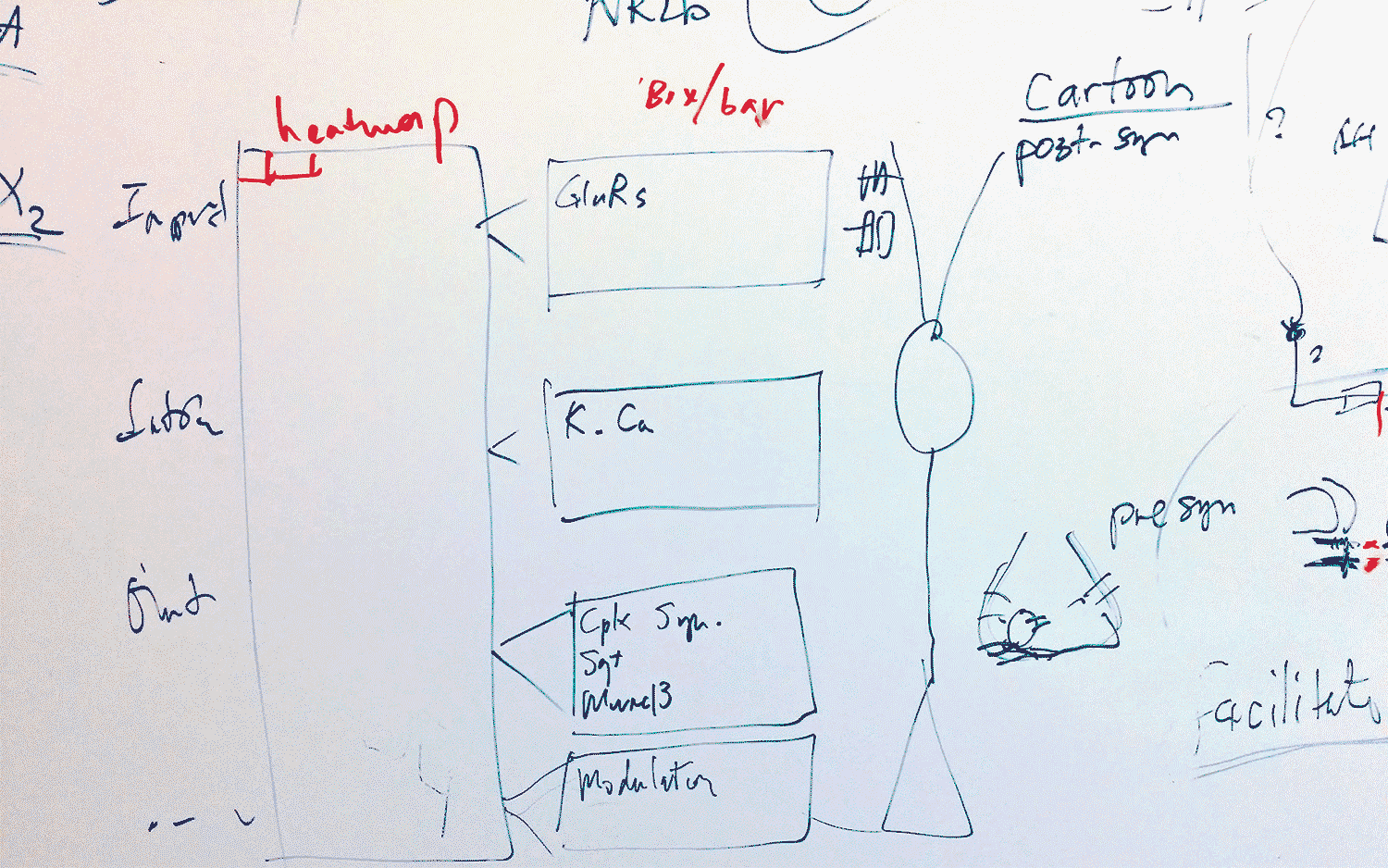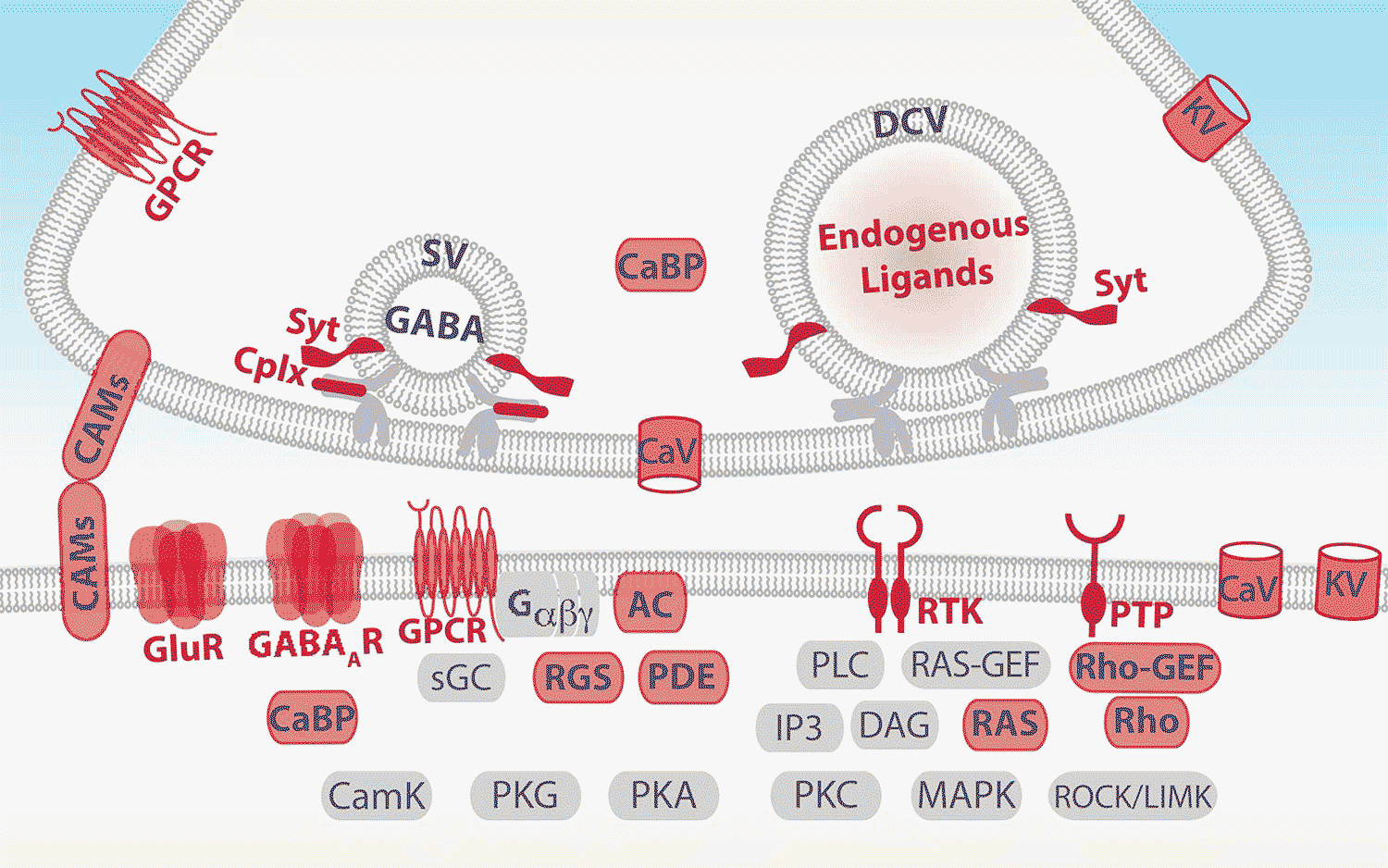
This is a discovery so fundamental it’s difficult to predict exactly where it will lead, but one aspect that Paul finds particularly exciting has to do with a class of communication equipment known as neuropeptides. It has long been known that neuropeptides are used in the brain to communicate messages between neurons that regulate basic activities like sleep and hunger. This research revealed “a huge cache” of neuropeptides within neurons involved in more complex tasks, including some implicated in mental illnesses, according to Paul.
“Now, that neuroscience is getting more refined in how we look into mood disorders, and autism, and schizophrenia,” he says “perhaps these neuropeptides are something that we have totally looked over.” Paul hopes that this discovery will help neuroscientists become more effective spies.