Gene Regulation and Inheritance
 The Gene Regulation and Inheritance Program focuses on revealing basic mechanisms governing the regulation of gene expression and cell inheritance at the DNA, RNA, and protein levels, and on discovering how these mechanisms are perturbed to influence the initiation and/or progression of cancer. A major strength of the Program is the innovative science that is yielding novel insights into non-coding RNA species, RNA splicing, chromatin biology, and cell-cycle control. Alterations in these processes are critical features of the transformed phenotype. Although fundamental research is the central to this Program, many discoveries are being translated toward the clinic, due in part to the strong strategic alliance with clinical partners.
The Gene Regulation and Inheritance Program focuses on revealing basic mechanisms governing the regulation of gene expression and cell inheritance at the DNA, RNA, and protein levels, and on discovering how these mechanisms are perturbed to influence the initiation and/or progression of cancer. A major strength of the Program is the innovative science that is yielding novel insights into non-coding RNA species, RNA splicing, chromatin biology, and cell-cycle control. Alterations in these processes are critical features of the transformed phenotype. Although fundamental research is the central to this Program, many discoveries are being translated toward the clinic, due in part to the strong strategic alliance with clinical partners.
Program Co-leaders
Adrian Krainer, Ph.D.
Justin Kinney, Ph.D.
![]() Members of the Gene Regulation and Inheritance Program share an interest in uncovering the mechanisms governing inheritance of cell state as well as mechanisms of transcriptional and post-transcriptional regulation, and on understanding how those mechanisms are altered in cancer cells. The Program has three main focus areas: (1) elucidating fundamental mechanisms governing the regulation of non-coding RNAs, transcription, and cell inheritance; (2) determining how transcriptional and post-transcriptional control are dysregulated in cancer; and (3) developing therapeutic agents and biological systems to target pro-tumorigenic alterations in transcriptional and post-transcriptional regulators. The Program also has expertise in computational analysis of gene expression patterns, mRNA splicing, and mutation identification which is being used to uncover alterations that drive aberrant gene regulation and impact all three focus areas. Program members combine cell, molecular biology, biochemical, structural biology, computational, and genetic approaches. The Program is enhanced by the excellent Cancer Center Shared Resources, especially the Animal, Sequencing Technologies & Analysis, Flow Cytometry, Microscopy, and Mass Spectrometry Shared Resources.
Members of the Gene Regulation and Inheritance Program share an interest in uncovering the mechanisms governing inheritance of cell state as well as mechanisms of transcriptional and post-transcriptional regulation, and on understanding how those mechanisms are altered in cancer cells. The Program has three main focus areas: (1) elucidating fundamental mechanisms governing the regulation of non-coding RNAs, transcription, and cell inheritance; (2) determining how transcriptional and post-transcriptional control are dysregulated in cancer; and (3) developing therapeutic agents and biological systems to target pro-tumorigenic alterations in transcriptional and post-transcriptional regulators. The Program also has expertise in computational analysis of gene expression patterns, mRNA splicing, and mutation identification which is being used to uncover alterations that drive aberrant gene regulation and impact all three focus areas. Program members combine cell, molecular biology, biochemical, structural biology, computational, and genetic approaches. The Program is enhanced by the excellent Cancer Center Shared Resources, especially the Animal, Sequencing Technologies & Analysis, Flow Cytometry, Microscopy, and Mass Spectrometry Shared Resources.
Nobel laureate honored at CSHL chemistry symposium
April 15, 2024
“The Future of Click Chemistry” brought together two-time Nobelist K. Barry Sharpless with his former apprentices John Moses and David Tuveson.
Up close and personal with cryo-EM
April 11, 2024
CSHL’s Cryo-Electron Microscopy course teaches the next generation of scientists to study life at the atomic level.
Click, click, boom—150 new molecules
April 4, 2024
CSHL Professor John Moses premieres an expansive line of new click chemistry products, uncovering leads for better antibiotics and cancer drugs.
CSHL’s Thomas Gingeras awarded $2 million NSF grant
April 3, 2024
Climate change threatens crops with acidic soils and aluminum toxicity. Gingeras leads an international team tackling this problem head-on.
From plant genomics to a bioscience revolution
March 25, 2024
CSHL played a lead role in mapping the first plant genome. Today, that breakthrough fuels a whole new understanding of life on Earth.
Cocktails & Chromosomes: Molecules to change the world
March 14, 2024
What is CSHL’s John Moses doing with that glowing liquid? Watch our expert chemist get a reaction from the crowd at Industry bar in Huntington, NY.
Why some RNA drugs work better than others
March 6, 2024
CSHL’s Justin Kinney and Spinraza inventor Adrian Krainer tested the newly approved SMA treatment, risdiplam, and another RNA therapeutic, branaplam.
Can AI uncover breast cancer risk factors?
February 26, 2024
This question lies at the heart of a new interdisciplinary collaboration between CSHL’s Camila dos Santos and Peter Koo.
How diet may impact cancer and possible treatments
February 1, 2024
Researchers at the CSHL Cancer Center study the links between disease and nutrition in hopes of uncovering new treatment and prevention strategies.
A quiz for the ages
January 29, 2024
Want to know the secret to a long life? So do CSHL scientists. Take this short quiz to see what they’ve found out about aging and longevity.
Joshua-Tor named CSHL Director of Research
January 2, 2024
The Cold Spring Harbor Laboratory professor and HHMI investigator steps into her new role effective January 2, 2024.
Dream big: A powerful vision for CSHL research
January 2, 2024
New Cold Spring Harbor Laboratory Director of Research Leemor Joshua-Tor shares her vision for the future of bioscience discovery.
CSHL celebrates 50th anniversary of McClintock Laboratory
November 20, 2023
Fifty years ago, CSHL honored Barbara McClintock by dedicating a building in her name. Today, it is home to four innovative cancer labs.
Breaking new ground: For science and society
November 13, 2023
CSHL’s Foundations for the Future campaign will propel the institution’s bioscience research and education programs to new heights and maximal impact.
Rob Martienssen awarded 2024 Genetics Society Medal
November 8, 2023
CSHL Professor Rob Martienssen earned the award for outstanding research contributions to the field of genetics.
First look: Whole-body gene expression
November 6, 2023
For the first time, scientists at CSHL have observed gene expression as it occurs throughout an animal. See life take shape in front of your eyes.
22nd Annual Women’s Partnership for Science Breaks Records
October 19, 2023
Cold Spring Harbor Laboratory held its 22nd Annual Women’s Partnership for Science lecture and luncheon.
Test your breast cancer awareness
October 18, 2023
Awareness is key to prevention and potential future treatments. Take this quiz to find out about the latest in breast cancer research at CSHL.
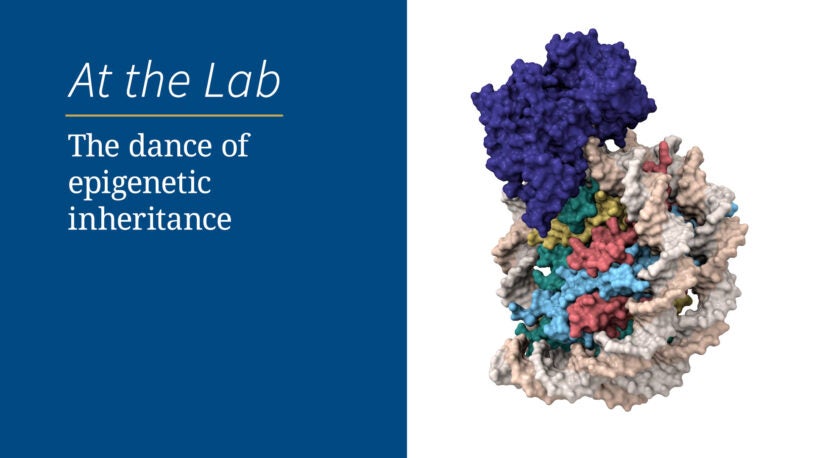
The dance of epigenetic inheritance
September 27, 2023
Making sure chromosomes get passed down correctly is hard work. Watch, through fluorescent and cryogenic lenses, how two proteins make it happen.
Stillman earns ASBMB Distinguished Scientist Award
September 7, 2023
The American Society for Biochemistry and Molecular Biology honored CSHL President & CEO Bruce Stillman for outstanding achievement in basic research.
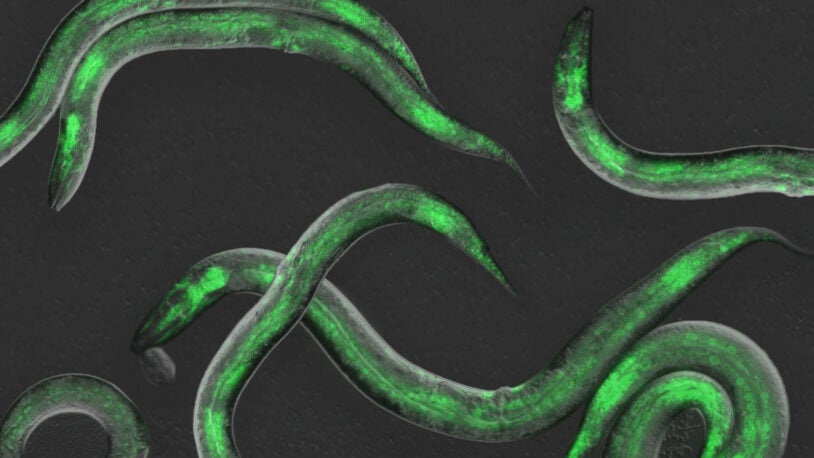
These worms have rhythm
September 5, 2023
Observing gene expression in real time, CSHL scientists identified four molecules the C. elegans worm relies on to set the tempo of its development.
How plants pass down genetic memories
August 28, 2023
Thirty years ago, CSHL’s Rob Martienssen discovered plant gene DDM1. Now, he’s identified just how the DDM1 protein helps control inheritance.
The ancient viruses in human DNA
August 24, 2023
CSHL Assistant Professor Andrea Schorn takes us into her lab for a behind-the-scenes look at the parts of our genome that aren’t quite human.
Where did our DNA come from?
August 22, 2023
Half the human genome isn’t quite human. CSHL’s Andrea Schorn gives us the inside scoop on how our DNA turned ancient viruses into essential allies.
CSHL makes a splash with Swim Across America
August 21, 2023
Since 1987, the charity swim has raised over $100 million for cancer research. Here, CSHL Assistant Professor Semir Beyaz voices his support.
Blending history and science
August 17, 2023
One of the most famous experiments conducted at CSHL relied on the same state-of-the-art equipment found in kitchens across the U.S. in 1952—a Waring blender.
Laying the groundwork for drug discoveries
August 8, 2023
A new partnership between CSHL and one of the world’s leading biotech investors could streamline this process and help change society for the better.
Eight serving one: CSHL volleyball mid-season report
August 2, 2023
CSHL’s 32nd Volleyball League season sees eight teams battling for the coveted Tiernan Cup and a year’s worth of bragging rights.
Bite into this diet and disease quiz
July 5, 2023
Test your knowledge of how diet and nutrition affect health and disease with this short quiz.
Adrian Krainer awarded honorary IADR membership
June 21, 2023
Krainer was recognized for his pioneering research on spinal muscular atrophy and RNA therapeutics.
Lingbo Zhang wins National Institutes of Health MERIT Award
June 15, 2023
The highly prestigious award will support Zhang’s research on the role of nutrients and other environmental factors in blood cancer development.
CSHL harnesses biology’s favorite chemical
June 7, 2023
Life on Earth depends on phosphorus to give DNA structure. Soon, biology’s chosen chemical could make for new cancer treatments and green materials.
The digital dark matter clouding AI
June 5, 2023
Scientists have unknowingly encountered mysterious noise while using AI to decipher our genetic code. CSHL has found a way to cut through the fog.
Summer 2023 Harbor Transcript now online
May 31, 2023
This Special Annual Report edition of CSHL’s magazine provides a look back at some of the Laboratory’s biggest stories from 2022.
President’s essay: Bringing bold visions to life
May 26, 2023
CSHL President & CEO Bruce Stillman sees the Laboratory as a global hub for scientific expertise and a powerful launchpad for early-career scientists.
The evolution of autism research
May 25, 2023
The conversation around autism has evolved over the past two decades. So has CSHL research. This retrospective shows how we’ve helped move the needle.
CSHL celebrates 20th graduating class
May 9, 2023
Friends, family, and faculty applauded 11 Ph.D. recipients at the CSHL School of Biological Sciences’ 2023 commencement.
The CSHL School of Biological Sciences’ class of 2023
May 7, 2023
The School of Biological Sciences awarded 11 Ph.D. degrees this year. Here, the graduates reflect on their time and experiences at CSHL.
AI training: A backward cat pic is still a cat pic
May 4, 2023
This basic rule of thumb is helping CSHL’s quantitative biologists train AI to get a better read of the human genome.
This killer protein causes pancreatic cancer
April 26, 2023
CSHL researchers have found that high levels of a protein called SRSF1 can cause pancreatitis and jumpstart tumor development.
Krainer named Society for RNA Therapeutics board member
April 18, 2023
He helped pioneer the field. It changed medicine. Now, he joins a group of renowned scientists and physicians aiming to take it to the next level.
From tragedy, a potential pediatric cancer treatment
April 12, 2023
CSHL Professor Adrian Krainer, the man behind the “miracle drug” known as Spinraza®, has found a way to fight a deadly pediatric brain cancer in mice.
New shape-shifting antibiotics could fight deadly infections
April 4, 2023
CSHL researchers have created a powerful new molecule that combats antibiotic-resistant superbugs.
A new, sustainable source for a promising cancer killer
March 23, 2023
This Malaysian jungle plant produces a chemical with remarkable anticancer properties. Now, CSHL scientists can synthesize that chemical in the lab.
A molecular machine’s secret weapon exposed
February 23, 2023
A shape-shifter with a protruding arm and an appetite for unwanted RNA! CSHL biochemists identify the hidden talents of a mysterious molecule.
Can you outsmart this AI quiz?
February 6, 2023
Think you’re plugged into the latest artificial intelligence advancements? Test your tech knowledge with this quiz on AI and computational biology.
PFF student named Regeneron scholar
January 30, 2023
CSHL Partners for the Future student Sean Krivitsky is a semifinalist in one the nation's most prestigious high school science competitions.
Cracking the mystery behind a deadly brain cancer
December 21, 2022
Scientists solve the mystery of how glioblastoma turns off cancer defenses without the usual cancer-inducing mutations.
CSHL breaks ground on new Neuroscience Research Complex
December 20, 2022
New York Lieutenant Governor Antonio Delgado announced a $30 million investment to help fund the new construction project.
The tiny plant tackling climate change
December 8, 2022
The humble aquatic duckweed plant has enormous potential as a new source of healthy protein, low-carbon biofuels, and other bioproducts.
Finding the right AI for you
December 5, 2022
AI’s popularity has reached a point where there are too many options. How do you know which AI is right for you? CSHL scientists have a solution.
Cancer lab makes surprise discoveries in heart disease
November 30, 2022
Two separate studies from the Spector lab at CSHL suggest that certain genes can lead to cardiac problems.
Welcome to Biology + Beyond
November 14, 2022
CSHL President and CEO Bruce Stillman introduces a special issue of Nautilus magazine now online, featuring the Lab’s latest groundbreaking research
A universal cancer treatment?
October 13, 2022
A medicine that disrupts the DNA replication of cancer cells may be within reach.
CSHL high schoolers finish top 10 in 2022 DREAM Challenge
October 7, 2022
The high school team competed against universities and private labs to build a computer program for predicting gene expression in yeast.
A new treatment approach for cystic fibrosis
July 14, 2022
Molecules called antisense oligonucleotides may help lung cells make a protein missing in people with cystic fibrosis.
The promising drug duo that may improve SMA treatment
July 11, 2022
Pairing Spinraza® with a second FDA-approved drug may be a new way to improve the drug’s therapeutic effects in spinal muscular atrophy patients.
Bruce Stillman honored with Excellence in Healthcare Award
June 27, 2022
The Long Island Herald recognized CSHL President and CEO Bruce Stillman for his leadership and impact in the biomedical field.
Krainer awarded Watanabe Prize in Translational Research
June 16, 2022
Indiana University School of Medicine honored Krainer for his pioneering work on RNA splicing, which led to the first FDA-approved SMA therapeutic.
Decoding how a protein on the move keeps cells healthy
May 31, 2022
The Argonaute protein is a workhorse for cell regulation and CSHL researchers discovered what helps it commute from job to job.
President’s essay: Foundations for the future
May 25, 2022
Strategically designed to spark scientific exchange and inspiration, CSHL is a unique research and education environment for advancing science.
CSHL Helix Society member honors late wife
May 24, 2022
Helix Society member John Broven recently visited CSHL to view a newly installed plaque placed in memory of his late wife.
CSHL 19th graduating class celebrated
May 2, 2022
In May 2022, the CSHL School of Biological Sciences awarded 10 Doctor of Philosophy degrees and two honorary degrees.
Cold Spring Harbor Laboratory 2022 Ph.D.’s
May 1, 2022
The School of Biological Sciences awarded Ph.D. degrees to ten students this year. Here are some stories and memories from their time at CSHL.
Martienssen elected to American Academy of Arts and Sciences
April 28, 2022
CSHL Professor and HHMI Investigator Rob Martienssen joins the American Academy of Arts and Sciences.
CSHL in pursuit of shape-shifting antibiotics
April 25, 2022
CSHL Professor John E. Moses was awarded over $325,000 from the New York State Biodefense Commercialization Fund to study a new type of antibiotic.
Can cancer be treated by changing its cells?
April 11, 2022
Tumors grow when cells lose their biological identity. A promising therapeutic might restore their sense of self.
Do you have the dirt on plant research?
March 31, 2022
New research is constantly sprouting. Take this quiz and test your plant knowledge.
Paving a path to triple-negative breast cancer treatment
March 29, 2022
Researchers collected a biobank of triple-negative breast cancer mini-tissues to search for new and potentially patient-specific treatments.
Regeneron competition honors CSHL high school researchers
March 22, 2022
Three high school student researchers at CSHL were among Regeneron Science Talent Search’s top 300 scholars. One made it to the final competition.
Stabilizing chromosomes to tackle tumors
March 17, 2022
Dicer and its partner BRD4 stabilize chromosomes. Targeting this pair could provide new therapeutic opportunities against cancer.
Tools of the trade at CSHL: Robotic microwave
February 24, 2022
This robotic assistant gives chemists a hand in the lab.
Cutting off liver cancer’s nutrient supply chain
February 3, 2022
CSHL researchers developed a way to interfere with the energy pathway that allows liver cancer to grow and spread.
Editing RNA to fix protein problems in cystic fibrosis
January 27, 2022
CSHL researchers found a new way to address a previously untreatable class of mutations in the cystic fibrosis gene.
The Darlene Carbone Brain Tumor Foundation donates $25,000 to CSHL
January 24, 2022
The Darlene Carbone Brain Tumor Foundation donates $25,000 to Dr. Alea Mills lab for glioblastoma research.
Lab meets clinic: Building on foundational research
December 15, 2021
The future depends on investments in scientific advancement, including expanding the Laboratory’s research visions and shoring up its infrastructure.
Teaching an old chemical new tricks
December 6, 2021
Using a chemical from the 1980s, CSHL Professor John E. Moses’ team has found a way to create new molecules in minutes.
In the Field: A Barbara McClintock–inspired novel
December 2, 2021
1983 Nobel laureate Barbara McClintock continues to inspire many today. In 2021, author Rachel Pastan published a novel based on her life and legacy.
Breaking the chain that culminates in cancer
December 1, 2021
CSHL scientists have discovered a way to shut down a cancer-causing protein by inhibiting a cascade of proteins that activate it.
Creating a community for hope
November 30, 2021
CSHL researchers began studying sarcoma in 2014, thanks in part to the encouragement and investments of three local foundations.
Unstoppable chemistry
November 5, 2021
“Click chemistry” is a way to design fast, specific, and clean reactions that make molecules click together like LEGO® bricks.
The “click” in click chemistry
November 4, 2021
Watch as CSHL Professor John E. Moses and Nobel laureate K. Barry Sharpless show click chemistry in action.
Tools of the trade at CSHL: NMR
October 29, 2021
Nuclear magnetic resonance—or NMR—uses magnetically generated radio waves to analyze chemical structures.
Masthead Cove Yacht Club supports CSHL research
October 20, 2021
The Masthead Cove Yacht Club raised $4,500 for CSHL research at their annual boat race.
Celebrating a new DNA Learning Center in Brooklyn
October 15, 2021
CSHL and CUNY opened a new DNA Learning Center in Brooklyn, NY. CSHL President & CEO Bruce Stillman explained the importance of genetics education.
The rise of RNA therapeutics
October 14, 2021
RNA has been making waves as a new approach to prevent or treat diseases, including COVID-19 and spinal muscular atrophy.
F.M. Kirby Foundation donates $115K for chemistry research
October 12, 2021
The F.M. Kirby Foundation donated $115,000 to support CSHL Professor John E. Moses’ chemistry research.
Building on 150 years of neuroanatomy
October 7, 2021
Learn more about how researchers reached a milestone in a years-long effort to catalog the cells of the human, mouse, and monkey brains.
Think a census of humans is hard? Try counting their brain cells!
October 6, 2021
CSHL researchers and other collaborators reached a milestone in a years-long effort to catalog the cells of the human, mouse, and monkey brains.
CSHL researcher wins NIH Director’s Pioneer Award
October 5, 2021
CSHL Adjunct Professor Z. Josh Huang was recognized for new cell engineering tools that will have broad applications in biological research.
Calculating the path of cancer
October 4, 2021
A new mathematical approach is helping cancer researchers at CSHL determine how mutations lead to different behaviors in cancerous cells.
Flagship DNA Learning Center NYC opens for all New Yorkers
September 24, 2021
Cold Spring Harbor Laboratory and the City University of New York announce the opening of the DNA Learning Center NYC at City Tech.
2021 Women’s Partnership luncheon raises $250,000
September 23, 2021
The twentieth annual Women’s Partnership for Science lecture and luncheon was held to support, promote, and celebrate women researchers at CSHL.
Krainer wins Pew grant to study fetal alcohol syndrome
September 15, 2021
CSHL Professor Adrian Krainer will study RNA splicing errors that occur in people with the disease and look for treatment targets.
CSHL President & CEO Bruce Stillman wins Advance Global Impact Award
September 8, 2021
CSHL President and CEO Bruce Stillman received the prestigious 2021 Australian Advance Global Impact Award.
CSHL Ph.D. program: Graduating class of 2021
August 22, 2021
The CSHL School of Biological Sciences awarded Ph.D. degrees to seven students this year, who describe some of their experiences.
Polymers “click” together using green chemistry
August 16, 2021
To build a new polymer using a type of green chemistry called “click chemistry,” chemists first had to tame a dangerous gas.
CSHL science tools at work: Rotovap
August 11, 2021
In the laboratory of John E. Moses, the rotating evaporator (rotovap) helps chemists purify the molecules they make.
CSHL serves up its 30th season of volleyball
August 3, 2021
People have been playing volleyball at CSHL for decades. The league returned for its 30th season in the summer of 2021.
URP: Summer camp for undergrads
July 29, 2021
The Undergraduate Research Program brings college students from around the world to CSHL for a summer of research and fun.
 Building publication list.
Building publication list.
Alexander Dobin
Next generation sequencing technologies revolutionized many areas of genetics and molecular biology, enabling quantitative analyses of the entire genomes and paving the way for Personalized Medicine. We develop novel statistical methods and computational algorithms for multi-omics processing and integration, and leverage Big Genomic Data to elucidate various problems in precision health, such as genetic and epigenetic mechanisms of cancer development and progression, and clinical impact of functional variants.

Thomas Gingeras
Only a small portion of the RNAs encoded in any genome are used to make proteins. My lab investigates what these noncoding RNAs (ncRNAs) do within and outside of cells, where regulators of their expression are located in the genome. This is particularly important in cancer. Our laboratory works on endometrial cancer and its relationship to age and obesity.

Christopher Hammell
As organisms develop, genes turn on and off with a precise order and timing, much like the order and duration of notes in a song. My group uses model organisms to understand the molecules that control the tempo of development. We also study how changes in the timing of gene expression contribute to diseases like cancer.

Leemor Joshua-Tor
Our cells depend on thousands of proteins and nucleic acids that function as tiny machines: molecules that build, fold, cut, destroy, and transport all of the molecules essential for life. My group is discovering how these molecular machines work, looking at interactions between individual atoms to understand how they activate gene expression, DNA replication, and small RNA biology.

Justin Kinney
Research in the Kinney Lab combines mathematical theory, machine learning, and experiments in an effort to illuminate how cells control their genes. These efforts are advancing the fundamental understanding of biology and biophysics, as well as accelerating the discovery of new treatments for cancer and other diseases.

Peter Koo
Deep learning has the potential to make a significant impact in basic biology and cancer, but a major challenge is understanding the reasons behind their predictions. My research develops methods to interpret this powerful class of black box models, with a goal of elucidating data-driven insights into the underlying mechanisms of sequence-function relationships.

Adrian R. Krainer
Our DNA carries the instructions to manufacture all the molecules needed by a cell. After each gene is copied from DNA into RNA, the RNA message is "spliced" - an editing process involving precise cutting and pasting. I am interested in how splicing normally works, how it is altered in genetic diseases and cancer, and how we can correct these defects for therapy.

Rob Martienssen
Chromosomes are covered with chemical modifications that help control gene expression. I study this secondary genetic code - the epigenome - and how it is guided by small mobile RNAs in plants and fission yeast. Our discoveries impact plant breeding and human health, and we use this and other genomic information to improve aquatic plants as a source of bioenergy.

David McCandlish
Some mutations are harmful but others are benign. How can we predict the effects of mutations, both singly and in combination? Using data from experiments that simultaneously measure the effects of thousands of mutations, I develop computational tools to predict the functional impact of mutations and apply these tools to problems in protein design, molecular evolution, and cancer.

Alea A. Mills
Cells employ stringent controls to ensure that genes are turned on and off at the correct time and place. Accurate gene expression relies on several levels of regulation, including how DNA and its associated molecules are packed together. I study the diseases arising from defects in these control systems, such as aging and cancer.

Lopa Mishra
My research focuses on the continuum of science-driven clinical care by working on novel therapies and improved clinical outcomes, honing liver disease, metabolism/alcohol, obesity/addiction gastrointestinal cancers, inflammatory bowel disease, and neural regulation of disease and cancer, which links to the field of bioelectronic medicine.

Andrea Schorn
Transposable elements make up half of our DNA. They control gene expression and have been a major evolutionary force in all organisms. The Schorn lab investigates how small RNAs identify and silence transposable elements when they become active during development and cancer.

David L. Spector
The immense amount of DNA, RNA and proteins that contribute to our genetic programs are precisely organized inside the cell's nucleus. My group studies how nuclear organization impacts gene regulation, and how misregulation of non-coding RNAs contributes to human diseases such as cancer.

Bruce Stillman
Every time a cell divides, it must accurately copy its DNA. With 3 billion “letters” in the human genome, this is no small task. My studies reveal the many steps and molecular actors involved, as well as how errors in DNA replication are involved in diseases that range from cancer to rare genetic disorders.

Lingbo Zhang
The research in the Zhang laboratory centers on normal and malignant stem and progenitor cells in the hematopoietic system and decodes the role of metabolites, including micronutrients and neurotransmitters, in the tumor microenvironment and their genetic effectors in regulating hematologic malignancies. The ultimate goal is to understand how environmental signals such as diets and nervous system activities modulate development and cancers.